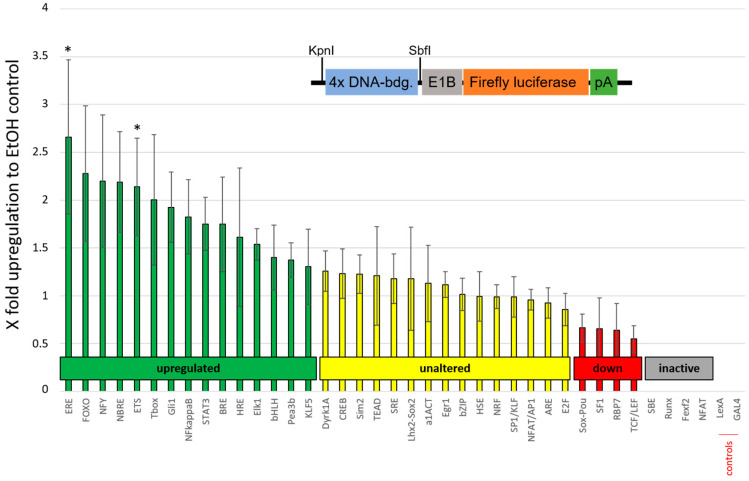
Figure 3.
Erinacine C differentially regulates reporter construct expression with binding sites for various transcription factors. Schematic drawing shows organization of Firefly luciferase reporter constructs containing a linear array of four consensus sequeces for transcription factor bindings sites (blue) followed by the weak basal promoter E1b (grey) and the cDNA coding for Firefly luciferase (orange) followed by the SV40 polyadenylation sequence (green). The sequences for the respective binding sites are provided in Table S1. Transcriptional activation of these constructs in 1321N1 cells upon incubation with erinacine C is displayed in relation to 0.5% EtOH solvent controls as fold-activation. Upregulated transcription above 1.3-fold over EtOH controls is shown in green color, while constructs with downregulated transcription below 0.7-fold with respect to EtOH controls are depicted in red color, these values for classifying and up- or downregulation were chosen arbitrarily for facilitating the readability of the data (four values per treatment, four independent cultures). * p < 0.05.