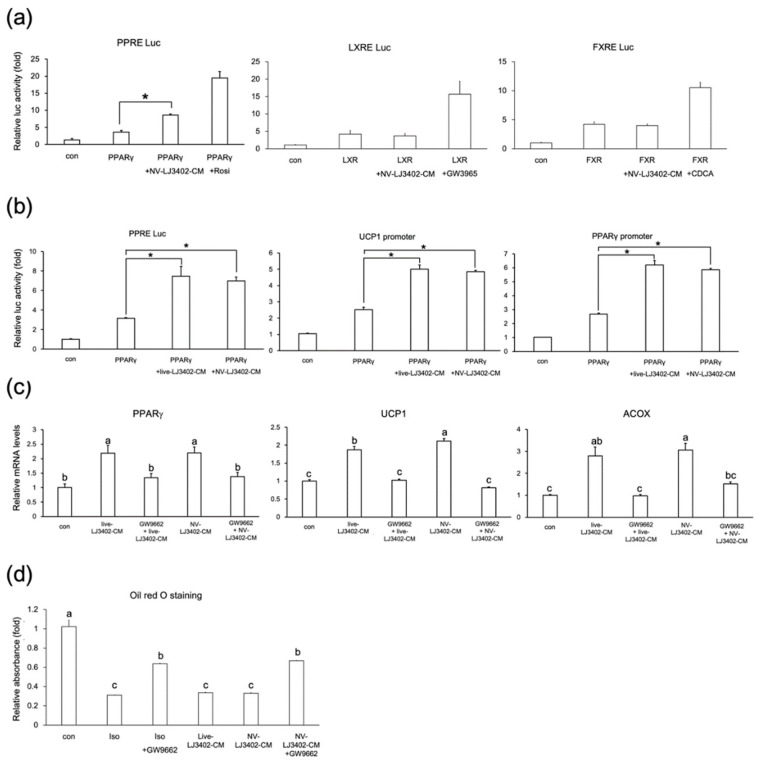
Figure 3.
Effect of NV-LJ3402 on the activities of transcription factors important for metabolic gene expression (a) HEK293T cells were co-transfected with different metabolic transcription factors (PPARγ, liver X receptor, LXR or farnesoid X receptor, FXR) and appropriate reporter genes harboring their cognate response elements (pGL3-ACOX-PPRE-Luc, PPRE Luc, pGL3-CETP-LXRE-luc, LXRE Luc or pGL3-TK-IR-1-Luc, FXRE Luc), as indicated. After 12-h transfection, cells were incubated for 24 h with either NV-LJ3402-CM, appropriate ligands, or vehicles (DMSO or con or both), as indicated, and then assayed for luciferase activity. (b) Effect of LJ3402-CM on PPARγ transcriptional activity was analyzed using various reporter genes harboring the PPARγ response element (pGL3-ACOX-PPRE-Luc) or different target gene promoters (pGL3-UCP1-Luc and pGL3-PPARγ-Luc) in HEK293T cells. After 12-h transfection, cells were treated with con, live LJ3402-CM, and NV-LJ3402-CM as indicated. (c,d) Day 6 3T3-L1 adipocytes were incubated with live LJ3402-CM, NV-LJ3402-CM, isoproterenol (Iso, 100 μM) or GW9662 (10 μM) or both for 48 h, as indicated. The mRNA levels (c) of genes coding for proteins involved in energy dissipation and lipid accumulation (d) were analyzed by RT-qPCR and Oil Red O staining, respectively, as indicated. All data are expressed as the mean ± S.E.M of three independent experiments. PPARγ vs. PPARγ+NV-LJ3402CM or live LJ3402-CM, * p < 0.05. Different lower-case letters above bars indicate significant differences (p < 0.05).