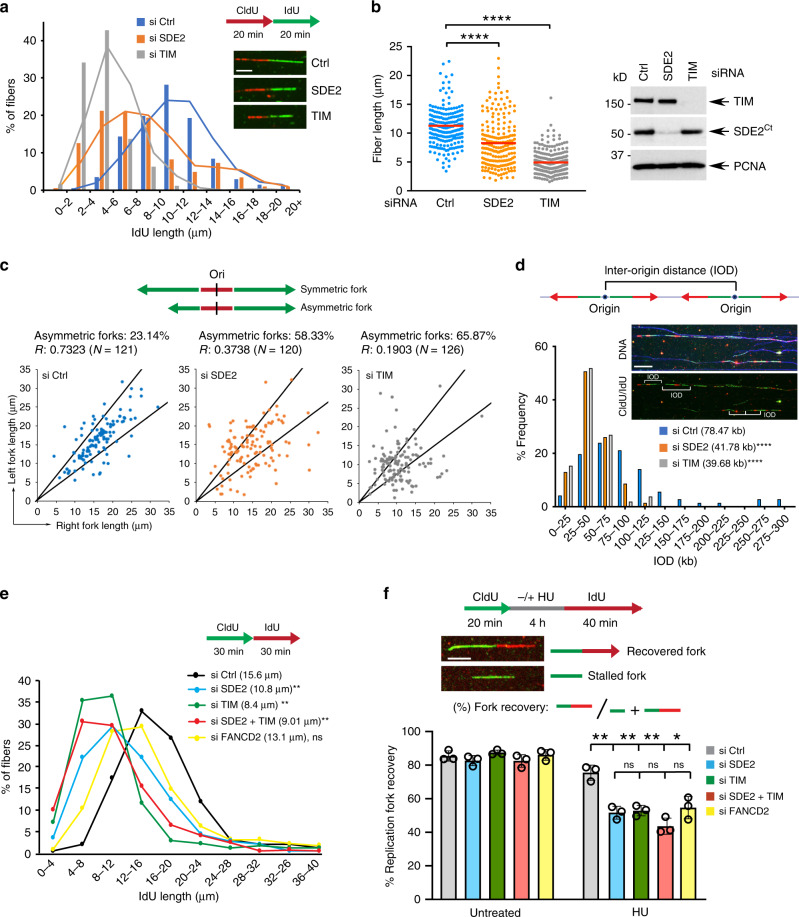
Fig. 4. SDE2 and TIM promote fork progression and stalled fork recovery.
a DNA combing analysis. Distributions of the IdU track length were determined on CldU:IdU double-labeled DNA fibers from siRNA-transfected U2OS cells (n = 2, a representative experiment is shown). b Left: dot plot of the IdU track length, n = 2 biologically independent experiments (****P < 0.0001, Mann–Whitney test). Red bars indicate the median value of at least 200 tracks per experimental condition. Scale bar, 5 μm. Right: WB to confirm knockdown. c Fork asymmetry analysis of siRNA-transfected U2OS cells. Sister forks from initiation events (green-red-green tracks only) are plotted from two technical replicates. The central area between the lines delimits a variation of <25% in sister forks length. R represents the linear correlation coefficient. d Distribution of inter-origin distance from siRNA-transfected U2OS cells. Upper panels show DNA counterstaining in blue and DNA tracks originated from CldU (green) and subsequently IdU (red) labeling (n = 2 biologically independent experiments, median value from at least 50 fibers, ****P < 0.0001, the two-tailed Mann–Whitney). Scale bar, 50 μm. e Distributions of the IdU track length from U2OS cells transfected with individual siRNA or in combination. FANCD2 knockdown, which does not impair fork progression, serves as a negative control. The median value of at least 200 tracks per experimental condition is indicated (n = 3, **P < 0.01, Student’s t-test). f Top: DNA combing protocol with a schematic of different replication dynamics. Fork recovery is measured as the portion of the number of green-red fiber tracks divided by that of green-only and green-red fibers. Scale bar, 5 μm. Bottom: replication fork restart after HU-mediated fork stalling in U2OS cells transfected with the indicated siRNAs (n = 3 biologically independent experiments, mean ± SD, **P < 0.01, *P < 0.05, Student’s t-test). FANCD2 knockdown serves as a control for a defect in fork recovery.