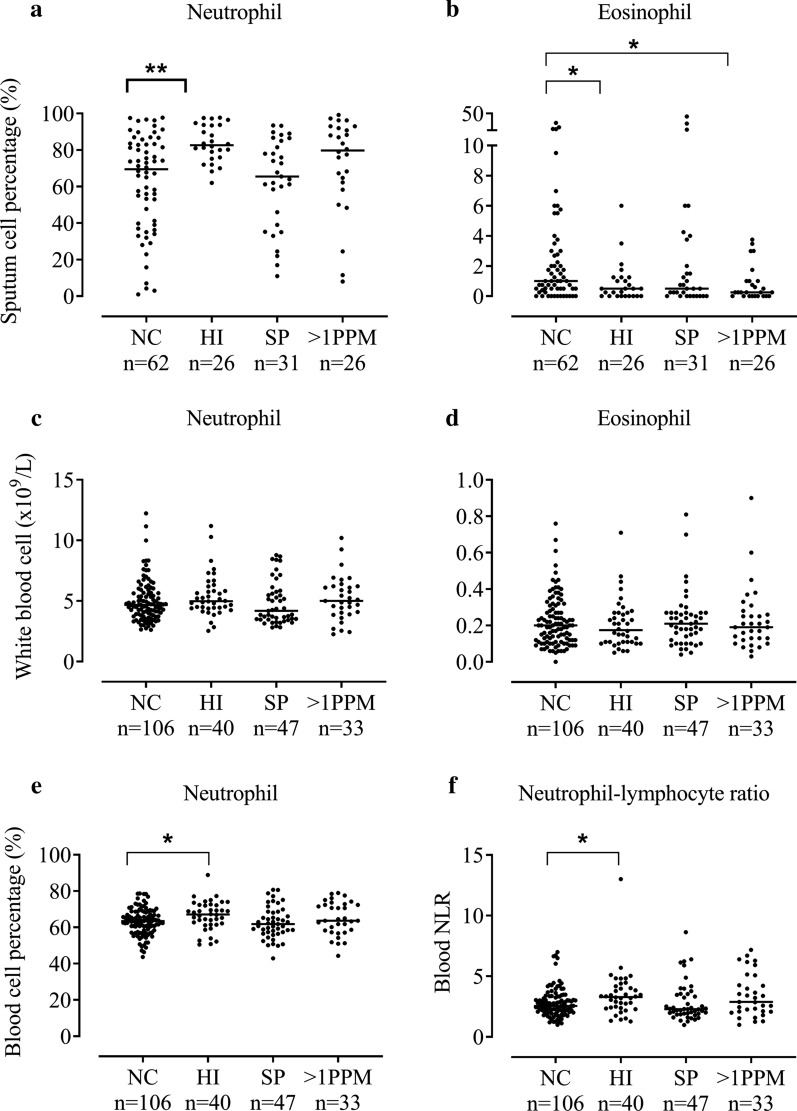
Fig. 1.
The comparison of sputum and blood eosinophil and neutrophil counts between patients colonised with different PPMs. Patients were categorised into four groups based on bacterial load defined by genome copies/ml of ≥ 1 × 104; no colonisation (NC), colonised with Haemophilus influenzae (HI), Streptococcus pneumoniae (SP) or > 1 potentially pathogenic microorganism (PPM). Sputum neutrophil % (n = 145) (a), sputum eosinophil % (n = 145) (b), blood neutrophil counts (n = 226) (c), blood eosinophil counts (n = 226) (d), blood neutrophil percentages (n = 226) (e) and blood neutrophil–lymphocyte ratio (n = 226) (f) are shown for each group. Statistical analysis was performed using Kruskal–Wallis and Mann–Whitney U adjusted for multiple comparisons. Data represent individual patients with median. *, **Significant difference to no colonisation group (p < 0.05, < 0.01 respectively)