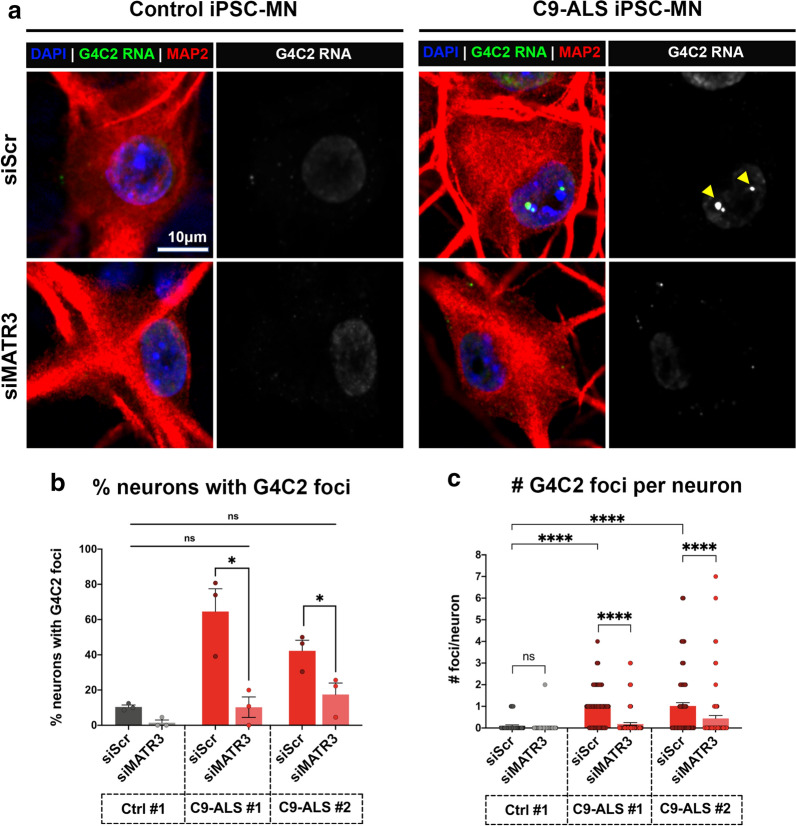
Fig. 6.
G4C2 RNA foci modulated by MATR3 in C9-ALS patient iPSC-MNs. a Representative confocal images of control (left) and C9-ALS patient (right) iPSC-MNs, indicated by MAP2 (red), transfected with scrambled siRNA or MATR3 siRNAs. FISH-IF was performed to stain nuclear G4C2 RNA foci (green; yellow arrowheads) in these neurons. b, c Quantification of the percentage of neurons that are G4C2 foci-positive (b), and number of G4C2 foci per neuron (c) in iPSC-MNs from one control (Ctrl #1) and two independent C9-ALS (C9-ALS #1, C9-ALS #2). Knocking down MATR3 significantly reduced the percentage of neurons that form G4C2 foci (b) and also reduced number of G4C2 foci per neuron (c) compared to scrambled-control in both C9-ALS iPSC-MNs (Kruskal–Wallis test). n = 24-50 neurons Each data point (circle) represents data from neurons pooled from 3 independent differentiations. Error bars indicate S.E.M. ****p value < 0.0001