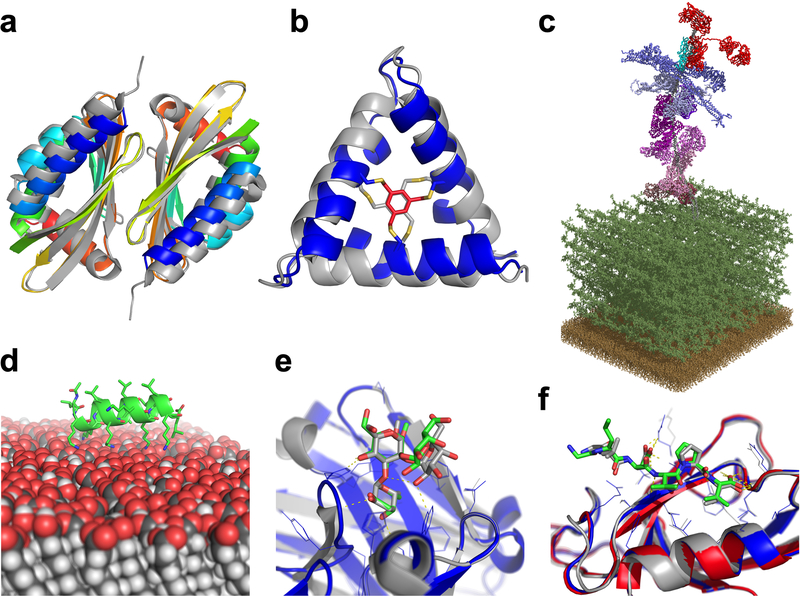
Figure 3: Rosetta can successfully address diverse biological questions.
(A) Curved β-sheet design: overlay of the designed homo-dimeric curved β-sheet (dcs-E_4_dim_cav3) in rainbow and the crystal structure in gray (PDBID 5u35). The protein is designed de novo and features a curved β-sheet, a large pocket, and a homodimer interface72. (B) Parametric design: overlay of the de novo designed macrocycle 3H1 in blue and the NMR structure in gray (PDBID 5v2g). This “CovCore” (covalent core) miniprotein is held together covalently by a hydrophobic cross-linker at its core (in red for the design and gray for the NMR structure)108. (C) PyTXMS: the interactome of M1 protein (virulence factor of Group A streptococcus) and 15 human plasma proteins on the surface of bacteria (peptidoglycan layer (dark green), and the membrane (brown)). This 1.8MDa structure contains over 200 chemical cross-links128 and is measured in a complex mixture of intact bacteria and human plasma. All models are provided by Rosetta: M1 protein (gray), IgG (red), four fibrinogens (dark to light blue), six albumins (dark to light pink), coagulation factor XIII A [F13A] (purple), C4bPa (cyan), haptoglobin [HP] (brown), and alpha-1-antitrypsin [SerpinA1] (plum). (D) RosettaSurface: model of an LK-α peptide (LKKLLKLLKKLLKL with a periodicity of 3.5 assuming a helical conformation) on a hydrophilic self-assembled monolayer surface. The peptide is unstructured in solution and assumes helical structure115 when on the surface, as experiments show. (E) RosettaCarbohydrate: flexible docking of a carbohydrate antigen to an antibody. The crystal structure is in gray (PDBID 1mfa) and the model in blue, with the carbohydrate in green. Antibody coordinates were taken from the PDB and glycan coordinates started from a randomized backbone conformation and rigid-body orientation143. (F) PIPER-FlexPepDock: high-resolution model of a peptide-protein complex (model: blue; solved structure in gray, PDBID 1mfg). The model was generated from a peptide sequence (LDVPV, derived from the C-terminal tail of ErbB2R) and the unbound structure of the receptor (Erbin PDZ domain, PDBID 2h3l, colored in red)111.