Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new strain of beta coronavirus that has spread worldwide within a short period of time and has been responsible for the current COVID-19 pandemic. This novel virus shows high transmission and adaptability frequency into the host with rapid changes in genomic sequences. In this study, we analyzed the complete genome of 41 strains isolated in Bangladesh to understand the evolutionary route and genetic variations of this rapidly evolving virus. The phylogenetics, parsimony informative sites and mutation analyses were performed using MEGA X, Multiple sequence alignment program (MAFFT), and Virus Pathogen Resource. The phylogenetic analysis of the studied genomes along with the reference genome suggested that the viral strains found in Bangladesh might be coming from multiple countries such as France, Germany, India, the USA, and Brazil. After entering into the country, intra-cluster and inter-cluster began to circulate in the 8 individual divisions of Bangladesh. We also identified 26 parsimony-informative sites along with the 9 most important sites for virus evolution. Genome-wide annotations revealed 256 mutations, of which 10 were novel (NSP3, RdRp, Spike) in Bangladeshi strains where I120F(NSP2), P323L(RdRp), D614G (Spike), R203K, G204R(N) are the most prominent. Most importantly, numerous mutations were flourishing in the N protein gene (67) followed by S (45), RdRp (38), NSP2 (34), NSP3 (20), and ORF8 (6) gene. Moreover, nucleotide deletion analysis found nine deletions throughout the genomes including in ORF7a (8), ORF8 (1) with one insertion (G) at 265 positions in only one genome. The underlying mechanism of disease severity, molecular evolution, and epidemiology lie in genomic sequences that are not fully understood yet. Identification of the evolutionary history, parsimony-informative sites and others genetic variations of this deadly virus will facilitate the development of new strategies to control the local transmission and provide deep insight in the identification of potential therapeutic targets for controlling COVID-19.
Keywords: Severe acute respiratory syndrome coronavirus 2, COVID-19, Bangladesh, Phylogenetic tree, Parsimony-informative sites
Highlights
-
•
Viral strains found in Bangladesh mainly came from multiple countries such as France, Germany, India, USA and Brazil.
-
•
Intra-cluster, and inter-cluster start to circulate in the 8 individual divisions of Bangladesh.
-
•
26 parsimony-informative sites along with 256 mutations were found in the circulating strains.
-
•
Nine deletions throughout the genomes including in ORF7a (8), ORF8 (1).
1. Introduction
The outbreak of a novel coronavirus (SARS-CoV-2) started from Wuhan city, China had an unprecedented impact within a short period of time around the world due to its highly contagious and spreading nature (Ahamed et al., 2020; Islam et al., 2020). According to recent data (6 August 2020) of the World Health Organization, this virus has already affected more than 215 countries and territories. As a result, more than 1,088,172 people have died around the world (https://www.worldometers.info/coronavirus/) due to this pandemic. In Bangladesh, since the first reported case of COVID-19 infection on 8th March 2020, 381,275 cases along with 5577 deaths have been reported as of 13th October 2020 (IEDCR, 2020). The first complete genome sequence of the virus in Bangladesh has been reported (EPI_ISL_437912) by the Child Health Research Foundation (CHRF) on the 15th of May 2020. Since then, more than 250 sequences have been published by several various other institutes in Bangladesh. From the genomic analysis, it has been shown that the size of the SARS-CoV-2 genome is approximately 29.8 kb to 29.9 kb (Rahaman et al., 2020; Saha et al., 2020a). This enveloped virus consists of polyprotein replicas (ORF1ab), and four structural proteins: spike protein (S), a membrane protein (M), an envelope protein (E), and nucleocapsid protein (N) (Rahman et al., 2020). There are also six accessory proteins encoded by ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10 predicted as hypothetical proteins in the SARS-CoV-2 genome (Wu et al., 2020; Rahaman et al., 2020). With the availability of complete genomes of SARS-CoV-2 strains, researchers are investigating the underlying mechanisms of severity, susceptibility hidden in the sequences (Khailany et al., 2020; Rahaman et al., 2020; Forster et al., 2020; Van Dorp et al., 2020). In this perspective, Bangladeshi researchers already have done numerous studies on Bangladeshi originated SARS-CoV-2 genome sequences (Ul Alam et al., 2020; Rahaman et al., 2020; Rahman et al., 2020; Hasan et al., 2020; Parvez et al., 2020; Saha et al., 2020b). Recently, an analysis conducted by Benvenuto et al. (2020) has revealed that viruses outside China formed a monophyletic clade, suggesting the effectiveness of border measures. In addition, another study by Li et al. in 2020 have revealed an high proportion (up to 25%) of genetic diversity among the studied China, Thailand and the United States originated SARS-CoV-2 genome. Several studies suggest that the virus developed mutations over a period of infection due to the antigenic drift process which allows them to change the characteristics (Coppee et al., 2020; Li et al., 2020). Along with all other genomic studies parsimony-informative sites analysis is very meaningful in the term of virus evaluation. Parsimony-informative is a site in where at least there is two different character states for the sequence at that site. This site contains at least two types of nucleotides (or amino acids), and at least two of them occur with a minimum frequency which is required for the fewest evolutionary changes in the genome. This informative positions are mainly helpful for giving information about evolutionary relationships among studied genomes. Moreover, in the evolutionary analysis this also important as it hypotheses relationships that require the smallest number of character changes is most likely to be correct (https://www.mun.ca/biology/scarr/2900_Parsimony_Analysis.htm). Moreover, studies on the SARS-CoV-2 genome have shown insightful information on phylogenetic relationships, mutational frequency, human-to-human transmission, and viral tropism (Lv et al., 2020; Ceraolo and Giorgi, 2020; Shang et al., 2020). This genome annotation along with parsimony-informative site analysis of SARS-CoV-2 might lead to improved understanding of transmission patterns, changes of behavior, and implementation of effective containment measures. In addition, the characterization of viral epidemiological nature, transmission root, and mutations can provide valuable information for deciphering the mechanisms linked to pathogenesis, immune evasion, and viral drug resistance. Therefore, Parsimony-informative sites, evaluation, and mutation studies can be crucial for the design of new vaccines, antiviral drugs and effective diagnostic tests. In this study, we aim to characterize the genetic variation and, parsimony-informative sites along with deletion and insertion site in circulating complete genomes of SARS-CoV-2 strains isolated in Bangladesh to assess the genetic diversity of SARS-CoV-2.
2. Materials and methods
2.1. Acquisition of SARS-CoV-2 genome sequences
Acquisition of SARS-CoV-2 genome sequences A total of 245 complete SARS-CoV-2 genome sequences of Bangladeshi origin, was submitted in the GISAID between 18 April and 30 July 2020 and were included in this study. Along with the complete genome sequences of 245 strains, with their date of collection and the patient's history, 35 reference genomes were selected based on the BLAST hit of the selected genomes (Supplementary Table 1). Partial genomes or genomes with unusually high variants including gaps or genome without a complete history of the patients/sampling place/collection date weren't considered in this study. Moreover, genome sequences containing with legionary characters (N, R, X, and Y) other than A, T, G, and C were also excluded from the analysis. Only complete genomes sequenced were used to compare with the reference genome (NC_045512.2).
2.2. Multiple sequence alignment
Sequence alignment was performed using a multiple sequence alignment program (MAFFT) command line (https://mafft.cbrc.jp/alignment/software/) (Katoh and Standley, 2013). Aligned sequences were finally opened with Molecular Evolutionary Genetics Analysis, across Computing Platforms (MEGA X) (Kumar et al., 2018) to remove all ambiguity and low-quality sequences.
2.3. Phylogenetic analysis of the retrieved Genome of SARS-CoV-2
After alignment and validation of all 245 retrieved genome sequences, the first 175 sequences were selected based on complete information and sequence quality. Finally, 41 out of 175 sequences with reference sequences were subjected to conduct phylogenetic analysis in maximum parsimony (MP) approach using MEGA X. In this MP method, the likelihood for each nucleotide substitution in the alignment was calculated (Kishino and Hasegawa, 1989).
2.4. Genomic variation analysis of SARS-CoV-2 sequences
Finally, the aligned sequences were visualized using MEGA X and Virus Pathogen Resource (https://www.viprbrc.org/) to identify the deletions and insertions with respect to the reference genome (NC_045512). Moreover, mutations that result in amino acid substitutions were investigated using the Wuhan reference sequence (NC_045512) where the GISAID platform mainly CoVserver enabled by GISAID in GIDAID EpiCoV data base along with a blast (https://blast.ncbi.nlm.nih.gov/) of all genome and separate proteins were used. The mutational frequency of the variants was determined where the ratio of the total number of mutations in each week and the total number of genomes obtained during that week were used to calculate the mutation frequency. Maximum Parsimony Tree and Parsimony informative sites were analyzed by using MEGA X.
3. Results
3.1. Genomic Analysis of SARS-CoV-2
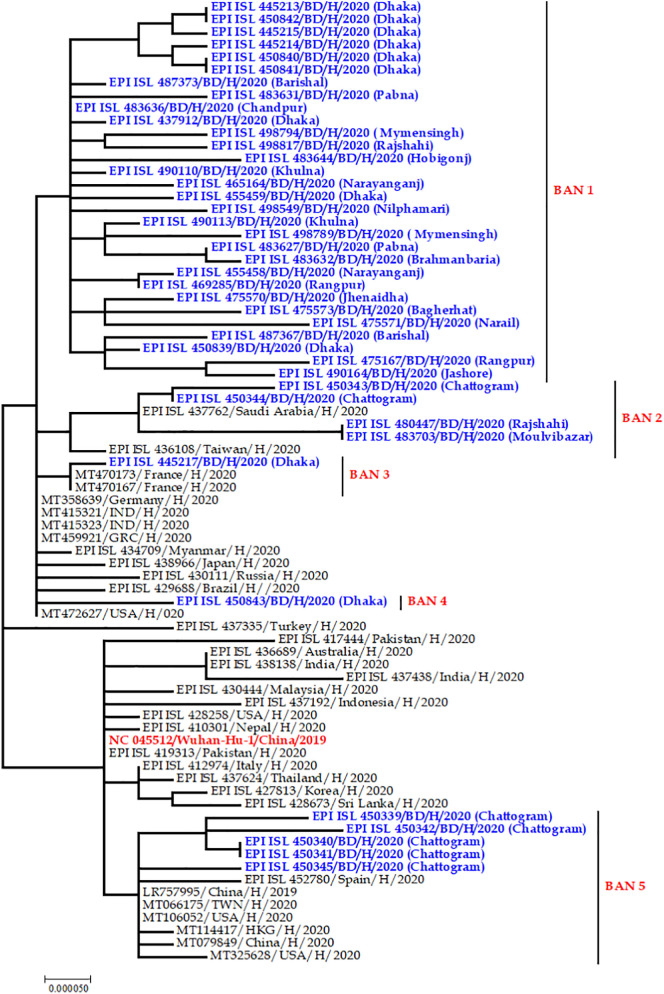
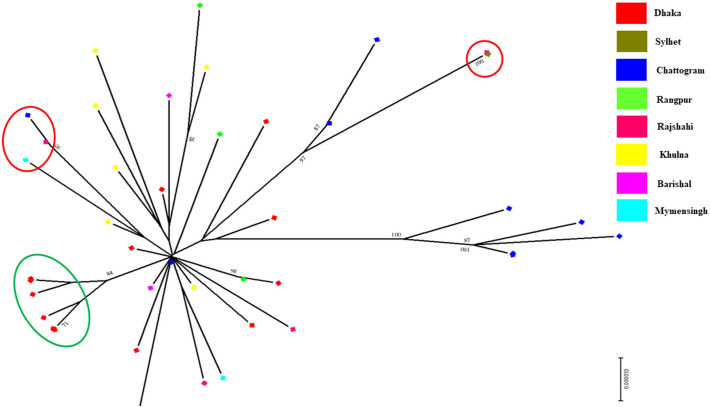
Initially, 70 of the 245 genome sequences were made redundant from the study and not analyzed due to it containing legionary characters and not meeting all the criteria of this study. After considering all the exclusion criteria, 175 unique SARS-CoV-2 genome sequence from eight different divisions (Dhaka, Sylhet, Chattogram, Rangpur, Rajshahi, Khulna, Barisal, and Mymenshing) in Bangladesh was investigated in this study (Supplementary Table 1). An MP phylogenetic tree was constructed in order to choose and identify potential and representing sequences from each and every reported division or area of Bangladesh (Supplementary Fig. 1). Finally, 41 representative genomic sequences from each clade (Supplementary Fig. 1) were considered for further analyses. During the representative genome selection process divisions and time of sequencing is also considered. The complete sequence of the 41 genomes with 35 reference genomes is provided in Supplementary file 1, where the accession numbers along with collection dates and place of all sequences are summarized in Supplementary Table 1. The tree also revealed the history of the common ancestry of all 41 genome sequences where the lines of a tree represent evolutionary lineages (Fig. 1 ). The tree also revealed that the Bangladeshi SARS-CoV-2 sequences differ from the reference sequence, NC_045512 (mark as red) of Wuhan, China followed by splitting into many sub-lineages, which are represented by different branches. Each branch of the tree ends with a cluster or a single sequence whereas closely related genome sequences were grouped in clusters (BAN 1 to BAN 5). For example, three Bangladeshi genome sequences were located in a BAN 2 group which originated in/ with genomes mainly from Saudi Arabia and Taiwan. Cluster BAN 3 and BAN 4 contain sequences were closely related to sequences from France, Germany, India, USA and, Brazil (Fig. 1). Moreover, five sequences of Cluster BAN 5 revealed mixed matching with the sequences from other countries of Asia, Europe and, Latin America. Most interestingly, group 1 (BAN 1) contained only sequences from Bangladeshi origin. We also found that some genome sequences, although isolated during the different months (April/May/June/July) and from different areas, were still closely related and therefore grouped in the same cluster (Fig. 2 ). From Fig. 2, we have found that in some cases a few circulating strains might cluster with the same strain circulating in this area which is indicated by the green circle in Fig. 2 where all isolates from Dhaka are clustering in the same clade. But intra distribution was also present (Sylhet and Rajshahi or Chattogram, Rajshahi, and Mymenshing isolate merged) which was indicated by the red line in Fig. 2. Our result also suggests that the circulating viral invading occurred independently or in clustery in every divisional region of Bangladesh (Fig. 3 ).
Fig. 1.
Phylogenetic analysis of 41 Bangladeshi SARS-CoV-2 complete genome sequences along with 35 sequences representing the Asian and rest of the world's sequences retrieved in this study. GISAID: Global Initiative on Sharing All Influenza Data; MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms; SARS-CoV-2: severe acute respiratory syndrome coronavirus. Main clusters are highlighted in blue colors. The Wuhan reference genome is in red color marked as reference sequence (GenBank accession number: NC_045512.2). Final log-likelihood was −38,467.62. Closely related genome sequences with minimum branch deviation (cut o 0.0005) were represented in clusters (BAN 1 to BAN 5). The viral genome sequences from Bangladesh are divided in to five clade marked as BAN 1 to BAN 5. Tree was built by using the best fitting substitution model (Kimura 2-parameter distance) through MEGA X software.
Fig. 2.
Phylogenetic connectivity analysis of 41 Bangladeshi SARS-CoV-2 complete genome sequences. GISAID: Global Initiative on Sharing All Influenza Data; MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms; SARS-CoV-2: severe acute respiratory syndrome coronavirus. The scale bar at the bottom of the tree represents 0.00010 nt substitutions per site. The representative sequences from all the reported area in Bangladesh were marked with diriment colors (Dhaka-red, Sylhet-Olive, Chattogram- Blue, Rangpur-green, Rajshahi- Faint red, Khulna- Yellow, Barisal- Pink, Mymenshing- Pest color). Red and green color circle indicating intra and inter clustering of the studied genome in the reported areas respectively.
Fig. 3.
Transmission of SARS-CoV-2 into Bangladesh showing possible multiple times entry into the Bangladesh from various Countries. Red dot arrow indicating the possible entry root. Red circle indicating the root of all the circulating virus in the Bangladesh whereas blue circle indicating the possible intervening countries used by the circulating genome during entry in to the Bangladesh. Image was taken from nextstrain, link https://nextstrain.org/ncov/global?f_country=Bangladesh.
3.2. Parsimony-informative sites analysis
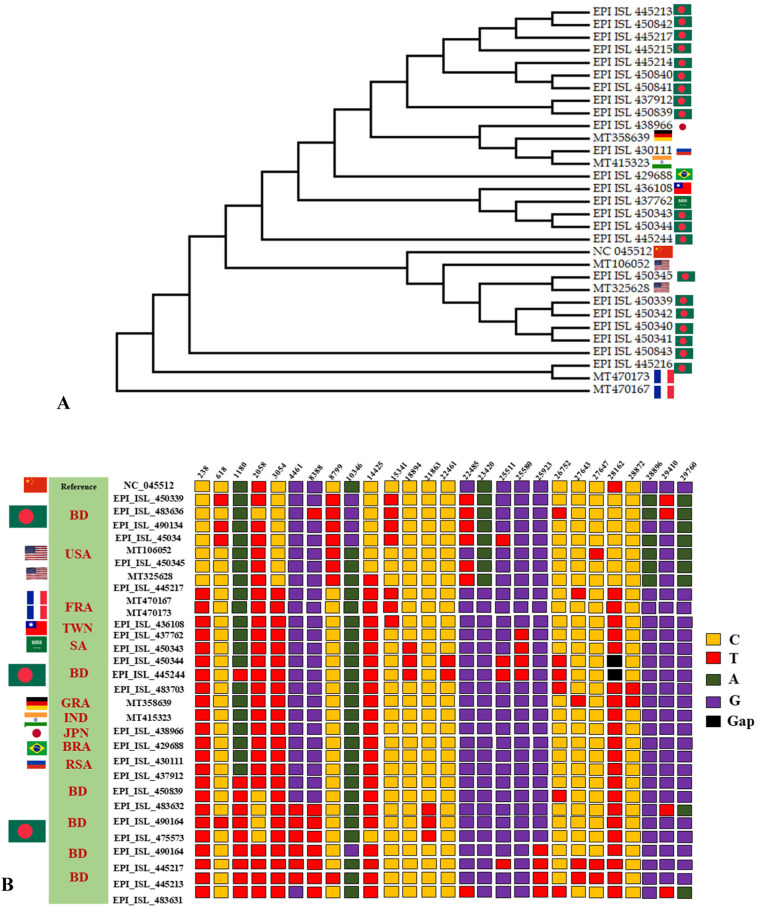
We found 26 parsimony-informative sites in which nine of them were important (Fig. 4 ) in conducting non-synonymous changes into the genome. The parsimony-informative sites were at 238, 618, 1180, 2058, 3054, 4461, 8388, 8799, 10346, 14425, 15241, 18894, 21863, 22461, 22485, 23420, 25511, 25580, 25923,26752,27643, 27647, 28162, 28872, 29410 and 29760 -th nucleotide of reference SARS-CoV-2 genome (NC_045512.2). The first important parsimony-informative site was 3054- nd nucleotide of referential SARS-CoV-2 genome (NC_045512.2) in which the referential nucleotide is C and the alternative nucleotide is T. The 7th important parsimony-informative site was 28872 -th nucleotide of the referential SARS-CoV-2 genome (NC_045512.2). The referential nucleotide is C and the alternative nucleotide is T. The remaining important parsimony-informative sites were 14425-th, 15341-th, 21863-th, 22461-th, 27643-th, 27647-th, and 28162 -th nucleotide covering changes of nucleotide C to T. Furthermore, the evolutionary history was inferred using the Maximum Parsimony method (Fig. 4). The Maximum parsimony analysis tree revealed that all the representative isolates from all five clusters might differ from each other. Moreover, some isolates may show less variance from others which are circulating in the same zone (Fig. 4 and Supplementary Table 1). In the tree the consistency index is 0.987500 (0.974359), the retention index is 0.993827 (0.983522), and the composite index is 0.981404 (0.968344) for all sites and parsimony-informative sites (in parentheses). However, this result clearly indicates that all the circulating viruses may continuously evolve and acquire synonymous or nonsynonymous changes into their genome and adapt themselves into the new environment (Dhaka-red, Sylhet-olive, Chattogram- blue, Rangpur-green, Rajshahi- faint red, Khulna- yellow, Barisal- pink, Mymenshing-pest color in Fig. 2).
Fig. 4.
Parsimony analysis of the circulating five group of Bangladeshi strain (Ban 1-5). A. Maximum parsimony analysis of representation isolates from 5 groups. The evolutionary history was inferred using the Maximum parsimony method. 1 out of 10 most parsimonious trees (length = 80) is shown. The consistency index is 0.987500 (0.974359), the retention index is 0.993827 (0.983522), and the composite index is 0.981404 (0.968344) for all sites and parsimony-informative sites (in parentheses). All positions containing gaps and missing data were eliminated. There were a total of 29287 positions in the final dataset. Evolutionary analyses were conducted in MEGA X. B. Parsimony-informative sites in representation isolates from 5 groups of SRAS-CoV-2 genome. After alignment, the SARS-Cov-2 sequences had 52 variant sites, of which 26 were parsimony-informative. The numerical value above the base indicates the position in the reference genome.
3.3. Variability analysis in SARS-CoV-2 Genome
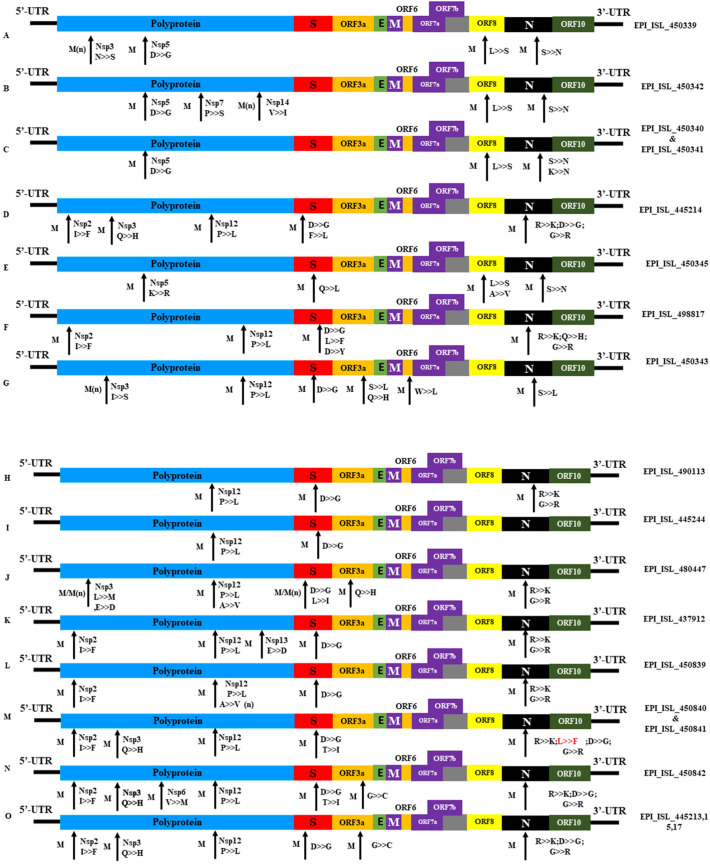
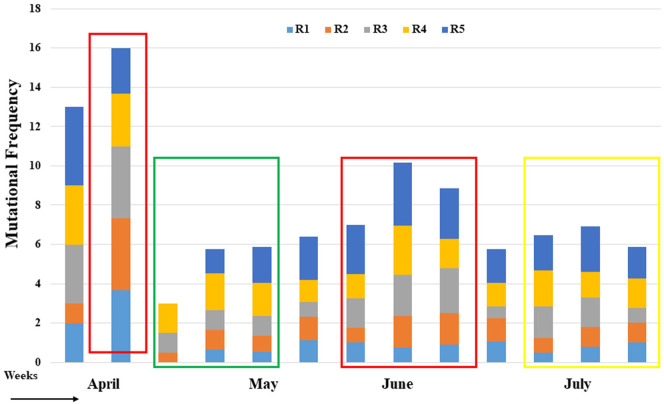
The sequence alignment analysis revealed a total of 256 mutations across the entire set of 41 genomes compared to the NCBI reference strain, Wuhan-Hu-1 (Accession NC_045512) (Supplementary Fig. 2 and Supplementary file 1). Among the 256 mutations, 10 are novel substitutions in NSP3, RdRp, and spike protein of the SARS-CoV-2 genome (Fig. 5 and Supplementary file 1). Among the identified nucleotide substitutions, 139 changes were found in the region of the structural protein (spike (S) glycoprotein-45; nucleocapsid (N) -67) followed by the 117 changes in polyprotein regions. In addition, the ORF3a, ORF6, ORF8 had a total of 14, 1, 6 nucleotide-level variations, respectively, whereas no nucleotide mutations were detected in 5′-UTR, 3′-UTR, and spacer regions (Fig. 5). In the ORF1ab polyprotein 38, 20, 34,6 aa substitutions have been identified in the NSP12, NSP3, NSP2, NSP5 respectively where's Helicase, 3′-to-5′ exonuclease, ORF7a, ORF7b all have only one aa substitutions. In case of spike protein, 34 predominant aa substitutions have been found in the 614 aa position (D>>G) followed by 30 and 29 aa substitutions at position 120 (I>>F) in NSP2 and 323 (P>>L) in RdRp respectively (Fig. 5 & Supplementary M file 1). On the other hand, aa substitutions at position 109 (F>>L) and 95(T>>L) were also found in the spike protein. We have observed two types of aa (H125Y, A2V) changes in the membrane (M) protein followed by no changes in the envelope (E) protein (Fig. 5). Besides the site-specific mutations, nine deletions (ORF7a = 8 & ORF8 = 1) of ranged nucleotides were found in 4 SARS-CoV-2 genome in Bangladesh (Fig. 5, Supplementary Fig. 3 & Supplementary M file 1). Furthermore, to identify the mutational frequency over time, the SARS-CoV-2 genome was divided into five regions (1 to 5) of approximately 6.5 kb each. The ratio of the number of total protein mutations along with the number of genome sequences in each week was considered during mutational frequencies calculation. Overall, highest mutational frequency was found in region 5 during the entire studied period followed by regions 4 and 2 (Fig. 6). Mutational frequency suddenly increased (red mark) mostly during the end of April but decreased again at the start of May (green mark). But most interestingly, at the beginning of June and mid-July (red mark) it increased again and at some points remained steady. But luckily at the end of July, this frequency was slowing down (green mark) (Fig. 6 and Supplementary M file 1). 183 & 35-nucleotide deletions were revealed in one studied viral strain (EPI_ISL_445213) ORF7a region followed by 49, 39, 35 -nucleotide deletion in EPI_ISL_445217 in the same region. In the strain, EPI_ISL_450842 3 deletion was found at 124, 70, 35 -nucleotide in ORF7a of the genome. Another noteworthy finding of our present systematic analysis is the identification of 344 nt deletions in ORF8 region in some circulating SARS-CoV-2 viral genome. Most interestingly one nt (G) insertion was also found in one SARS-CoV-2 genome in Bangladesh at 265 nt position in NSP1 (Supplementary Fig. 3).
Fig. 5.
Genomic mutation analysis of SARS-CoV-2. Genomic mutation analysis of SARS-CoV-2 strains identified. A-O marking indicating the mutational changes among representative Bangladeshi SARS-CoV-2 strain studied in this study in comparison with the reference strain NC_045512.2. Here, M-mutation; M(n)- Novel mutation, Protein: G- glycine, L-Leucine, I- isoleucine, P-Proline, Y-Tyrosine, W-Tryptophan, S-Serine, T-Threonine, C-Cysteine, M-Methionine, N-Asparagine, Q- Glutamine, D-Aspartate, K-Lysine, R-Arginine, H-Histidine.
Fig. 6.
Mutational frequency in circulating SARS-CoV-2 genome in Bangladesh between April to July. Mutational frequency was calculated by the ratio of the number of total protein mutations and the number of genome sequences in each week. The SARS-CoV-2 genome was divided into five regions, which are represented as R1–R5.Red (High), green (Low) and yellow (Moderate) box indicating the all possible fluctuations of the mutational frequency in all circulating SARS-CoV-2 genome in Bangladesh.R1- Leader, NSP2; R2- NSP3-9; R3- NSP10, NSP11, RDRP, Helicase, 3′-to-5′ exonuclease; R4- EndoRNase, 2′-O-ribose methyl transferase, Spike, ORF3a; R5- Envelope, Matrix, ORF6, ORF7a, ORF7b, Nucleocapsid,ORF8.
4. Discussion
The genetic information of SARS-CoV-2 is protected in its genome as like as other life, and annotation is the initial step to interpret the sequence. As a consequence of it, more than 103,770 genome sequences of SARS-CoV-2 has been screened around the world to reveal the hidden footprint in to the circulating genome and find the easiest possible solution to stop the ongoing deadly pandemic as soon as possible. Unfortunately till now there is no available vaccine against COVID-19 and the high error rate during RNA synthesis provides RNA genomes high mutation rates (Duffy, 2018). As a result of a higher mutation rate, effective vaccine design against RNA virus needs a close genomic analysis of all circulating strains. In this study, we have investigated the molecular variation between the recently sequenced Bangladeshi originated genomes of SARS-CoV-2 in comparison with relevant sequences reported throughout the world to know the exact origin of the circulating strains and their genotypic variation. A total of 41 from 175 samples were analyzed in the study. Phylogenetic analysis of the circulating strains revealed 5 major clades that share close sequence similarity with isolates from the United States, Saudi Arabia, France, Russia, India, and Germany. This result suggests that there is a high possibility the virus might have reached in Bangladesh from one of the European countries like Italy, Spain, France, and Germany. In a similar study conducted by Rebecca et al. in 2020 showed that in the early stages of the COVID-19 pandemic, overseas-acquired infections dominated in Australia with multiple lineages of SARS-CoV-2 from regions with high transmission rates such as Asia, Western Europe, and North America. In another study by Hasan et al. in 2020, on Bangladeshi 1st reported strain revealed that the virus might have reached Bangladesh from Arizona, the United States. But due to the continuous transportation and lack of stick rules, exchanging various circulating genomic clusters is also found in Bangladesh (Fig. 2). Various circulating genomic clusters are also found in the strains, possibly due to the continuous transportation and lack of stick rules. The parsimony-informative analysis showed 26 parsimony-informative sites with nine important regions in the virus evolves. Similarly, another study conducted by Matsuda et al. (2020) reported that viruses that invaded Taiwan, the United States, and Japan were introduced independently by having thirteen parsimony-informative sites that played an important role in the evolution of the virus in those countries. In our study we have found that Bangladeshi isolates fell into clade A2a and Clade B4 and contains 256 mutations with 10 novel mutations. Among the mutations, the most frequent 2 mutations were D614G (Spike protein) and P323L (NSP12). Moreover, D614G substitution may affect the structure of spike glycoprotein which helps the virus to enter into the host cell by binding to the ACE2 receptor. Virus virulence, transmission, and pathogenicity might be greatly affected by the reported unique mutations in NSP3 followed by S, RdRp protein in this study. In addition, Adam Brufsky et al. in 2020 suggested that heavy alteration in the glycosylated spike protein might be resulted due to the D614G mutation. This alteration affect the membrane fusion in tissues resulting in a change of the virulence pathogenic properties (Brufsky, 2020). However, a recent study in India by Sarkar et al. in 2020 identified a A930V mutation in the S glycoprotein, which was absent in our sequences. Besides the site-specific mutations, nine deletions of ranged nucleotides were found in polyprotein ORF7a = 8 & ORF8 = 1 in 5 strains. These nucleotide deletions can influence potentially the tertiary structures and functions of the polyprotein, S, M, and E proteins which may play an important role in virus-host interactions for infections, pathogenesis as well as immuno-modulations (Li et al., 2020; Xu et al., 2020; Zhou et al., 2020). Furthermore, several deletion mutations in ORF7a and ORF8 have also been observed in the analysis which have greater impact on viral host adaptation followed by transmission and replication (Su et al., 2020). A recent study by Holland et al. (2020) has also found this type of mutation in the circulating strain in Arizona which is also supported by another study conducted by Su et al. in 2020 and Parvez et al. in 2020. In addition, the 382-nucleotide deletion was also reported spanning from downstream of ORF7b to the ORF8 region by Liu et al. in 2020 which have an impact of enhanced transcription of the subsequent N protein (Su et al., 2020). Moreover, Nelson et al. in 2005 have presented immunoprecipitation data in case of SARS, which suggests an interaction between ORF7a and ORF8 with proteins M, E, and S (Nelson, 2005). They stated that ORF7a, ORF8 is important for viral assembly, viral replication cycle. Hence, more detailed experiments are needed to determine the functional consequences of the ORF7a and ORF8 deletion.
Notably, Liu et al. in 2020 identified two common deletions at the upstream of the polybasic cleavage site of S1-S2 (Liu, 2020). However, no such deletion was observed in our study. In another study by Khailany et al. (2020), the most common variants were reported in ORF1ab (C8782T), ORF8 (T28144C) and N gene (C29095T). Moreover, in the case of RT-PCR-based SARS-CoV-2 diagnostic generally RdRp, E, and N genes are targeted by various researchers (Khailany et al., 2020), due to their high sequence conservation. So diagnosis of the SARS-CoV-2 might be hampered by any kind of influential changes in targeted genes (Khailany et al., 2020). In our study among all the studied genomic diagnostic assay, 5 sets of genomic diagnostic assay contain the mismatches in the targeted genomic portion which is partially supported by the study of Khan and Cheung (2020) who reported mismatches in 7 sets of genomic diagnostic assay out of 27 studied assays (Supplementary Table 2) which could affect the diagnostic procedure in Bangladesh. Therefore, mutation profiles of the circulating SARS-CoV-2 isolates in the country should be taken into account.
As SARS-CoV-2 genomes spread, they leave footprints behind (mutations) allowing us to trace them. From this footprint, we have found that mutational frequency in the circulating SARS-CoV-2 genome in Bangladesh is fluctuating over time which is supported by another recent study by Saha et al., 2020c. It is feasible to complement the conventional approach with genome sequencing in an unbiased way. This fight against ongoing deadly COVID-19 pandemic will be a long one until we develop an effective treatments.
The following are the supplementary data related to this article.
Genomic Details of the all studied Bangladeshi originated SARS-CoV-2 genome.
Genomic view along with meta data of all studies SARS-CoV-2 Genome sequences in Bangladesh.
Funding source
No funding
CRediT authorship contribution statement
O.S. developed the hypothesis, drafted and reviewed the manuscript, M.H.H. reviewed the manuscript, M.M.R. supervised the whole work and critically reviewed the drafted manuscript. All authors read and approved the final manuscript.
Declaration of completing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Bangabandhu Science & Technology Fellowship Trust for supporting Otun Saha with PhD fellowship.
References
- Ahamed M.M., Naznin R.N., Saha O., Rahaman M.M. Recommendation of fecal specimen for routine molecular detection of SARS-CoV-2 and for COVID-19 discharge criteria. Pathog. Glob. Health. 2020;1 doi: 10.1080/20477724.2020.1765651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D., Giovanetti M., Salemi M., Prosperi M., De Flora C., Junior Alcantara L.C. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathog. Glob. Health. 2020:1–4. doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brufsky A. Distinct viral clades of SARS-CoV-2: implications for modeling of viral spread. J. Med. Virol. 2020;92:1386–1390. doi: 10.1002/jmv.25902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92(5):522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppee F., Lechien J.R., Decleves A.E., Tafforeau L., Saussez S. Severe acute respiratory syndrome coronavirus 2: virus mutations in specific European populations. New Microbes New Infect. 2020;36:100696. doi: 10.1016/j.nmni.2020.100696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. 2020;117(17):9241. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S., Khan S., Ahsan G.U., Hossain M.M. 2020. Genome Analysis of SARS-CoV-2 Isolate from Bangladesh. (BioRxiv) [Google Scholar]
- Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., Lim E.S. An 81 nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (Jan-Mar 2020) J. Virol. 2020 doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Epidemiology, Disease Control and Research COVID-19 Status Bangladesh. https://www.iedcr.gov.bd/ Available from.
- Islam A. Challenges to be considered to evaluate the COVID-19 preparedness and outcome in Bangladesh. Int. J. of Healthc. Manag. 2020:1–2. [Google Scholar]
- Katoh Kazutaka, Standley Daron M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K.A., Cheung P. Presence of mismatches between diagnostic PCR assays and coronavirus SARS-CoV-2 genome. R. Soc. Open Sci. 2020;7(6) doi: 10.1098/rsos.200636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino Hirohisa, Hasegawa Masami. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989;29(2):170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Xingguang, Wang Wei, Zhao Xiaofang, Zai Junjie, Li Yi, Chaillon Antoine. Transmission dynamics and evolutionary history of 2019‐nCoV. J. Med. Virol. 2020;92(5):501–510. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Zhe. Identification of Common Deletions in the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020;94(17):1–9. doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Li G., Chen J., Liang X., Li Y. 2020. Comparative Genomic Analysis Revealed Specific Mutation Pattern between Human Coronavirus SARS-CoV-2 and Bat-SARSr-CoV RaTG13. (BioRxiv) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Suzuki H., Ogata N. 2020. Phylogenetic Analyses of the Severe Acute Respiratory Syndrome Coronavirus 2 Reflected the Several Routes of Invasion in Taiwan, the United States, and Japan. arXiv Preprint arXiv. [Google Scholar]
- Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13(1):75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez M.S.A., Rahman M.M., Morshed M.N., Rahman D., Anwar S., Hosen M.J. 2020. Genetic Analysis of SARS-CoV-2 Isolates Collected from Bangladesh: Insights into the Origin, Mutation Spectrum, and Possible Pathomechanism. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman Md Mizanur, Saha Otun, Rakhi Nadira Naznin, Chowdhury Md Miraj Kobad, Sammonds Peter, Kamal A.S.M. Maksud. Overlapping of locust swarms with COVID-19 pandemic: a cascading disaster for Africa. Pathog. Glob. Health. 2020;114(6):285–286. doi: 10.1080/20477724.2020.1793595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.S., Hoque M.N., Islam M.R., Akter S., Rubayet-Ul-Alam A.S.M., Siddique M.A., Hossain M.A. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: an in silico approach. PeerJ. 2020;8:9572. doi: 10.7717/peerj.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha Otun, Rakhi Nadira Naznin, Towhid Syeda Tasneem, Rahaman Md Mizanur. Reactivation of severe acute respiratory Coronavirus-2 (SARS-CoV-2): hoax or hurdle? Int. J/Healthc. Manag. 2020:1–2. [Google Scholar]
- Saha O., Shatadru R.N., Rakhi N.N., Islam I., Hossain M.S., Rahaman M.M. 2020. Temporal Landscape of Mutation Accumulation in SARS-CoV-2 Genomes from Bangladesh: Possible Implications from the Ongoing Outbreak in Bangladesh. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Malaker R., Sajib M.S.I., Hasanuzzaman M., Rahman H., Ahmed Z.B.…Vanaerschot M. Complete Genome Sequence of a Novel Coronavirus (SARS-CoV-2) Isolate from Bangladesh. Microbiology Resource Announcements. 2020;9(24) doi: 10.1128/MRA.00568-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.C., Anderson D.E. 2020. Discovery of a 382 -nt Deletion during the Early Evolution of SARS -CoV -2. bioRxiv. [DOI] [Google Scholar]
- Ul Alam A.R., Rafiul Islam M., Shaminur Rahman M., Islam O.K., Anwar Hossain M. Understanding the possible origin and genotyping of first Bangladeshi SARS-CoV-2 strain. J. Med. Virol. 2020;1–4 doi: 10.1002/jmv.26115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Ortiz A.T. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus 396 associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;1-4 doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nat. 2020:1–4. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic Details of the all studied Bangladeshi originated SARS-CoV-2 genome.
Genomic view along with meta data of all studies SARS-CoV-2 Genome sequences in Bangladesh.