Abstract
The health of mammals depends on a complex interplay with their microbial ecosystems. Compartments exposed to external environments such as the mucosal surfaces of the gastrointestinal tract accommodate the gut microbiota, composed by a wide range of bacteria. The gut microbiome confers benefits to the host, including expansion of metabolic potential and the development of an immune system that can robustly protect from external and internal insults. The cooperation between gut microbiome and host is enabled in part by the formation of partitioned niches that harbor diverse bacterial phyla. Bacterial secretion systems are commonly employed to manipulate the composition of these local environments. Here, we explore the roles of the bacterial type VI secretion system (T6SS), present in ~25% of gram-negative bacteria, including many symbionts, in the establishment and perturbation of bacterial commensalism, and symbiosis in host mucosal sites. This versatile apparatus drives bacterial competition, although in some cases can also interfere directly with host cells and facilitate nutrient acquisition. In addition, some bacterial pathogens cause disease when their T6SS leads to dysbiosis and subverts host immune responses in defined animal models. This review explores our knowledge of the T6SS in the context of the “host-microbiota-pathogen” triumvirate and examines contexts in which the importance of this secretion system may be underappreciated.
Keywords: gut microbiome, type six secretion system, commensal, symbiosis, dysbiosis, mucosal immunity, tolerance, MAMPs
Introduction
The gut tissue is composed of hundreds of millions of cells whilst providing a home for a microbiota containing trillions of bacteria (Sender et al., 2016). The association of the microbiota with our tissues is central for homeostatic and developmental mechanisms and thus governs many aspects of human health (Belkaid and Harrison, 2017). Due to this relationship, mammals in general may be considered as holobionts from an ecological perspective, in which the microbiota assists host metabolism and acts as an environmental training system for the associated tissues (Bäckhed et al., 2005; Al Nabhani et al., 2019; Tsolis and Bäumler, 2020). Microorganisms associate with the skin and mucosal surfaces such as the oral-nasal and vaginal cavities, respiratory and gastrointestinal tracts; with the gut microbiota constituting the best characterized community. We note that although we focus on the gut microbial ecosystem, the concepts may apply to all mucosal surfaces and potentially to other complex symbiotic communities.
The composition and community structure of the gut microbiota is complex and heterogenous. The distribution of microbial species within the large intestine is to be accounted with the diversity of residing immune cells, together forming a biodynamic ecosystem (Human Microbiome Project Consortium, 2012; James et al., 2020). Indeed, bacterial communities and immune cell populations exhibit great diversity in a niche-dependent fashion, with the latter displaying a wide range of transcriptional profiles within T and B cells of the adaptive immune system. The niches of gut commensals are determined by their metabolic activities and ability to stably associate with their local tissue environment (Lee et al., 2013; Ost and Round, 2018; Vonaesch et al., 2018). For example, some of the Bacteroides species are present in the intestinal lumen while others tightly associate with the mucus layering the epithelial surface of colonic crypts (Johansson et al., 2011). Yet, niche residency is not solely determined through dialogue with the host and critically depends on interactions with other microbes sharing nutritional niches (García-Bayona and Comstock, 2018). Here, bacteria vie for dominance, deploying a range of antibacterial toxins, some of which are delivered via membrane-embedded secretion systems.
The T6SS is prevalent in gram-negative bacteria, particularly in the phyla Proteobacteria and Bacteroidetes (Bingle et al., 2008; Russell et al., 2014b). This secretion apparatus is evolutionarily related to the bacteriophage tail, wherein contraction of a sheath propels a spiked-tube structure out of the bacterial cell, piercing the cell membrane of their targets to inject effector proteins (Pukatzki et al., 2007; Coulthurst, 2019). The cytoplasmic T6SS sheath, composed of a polymeric helix of TssB-TssC binds to a baseplate-like multi-protein platform, which itself associates with an envelope-spanning membrane complex of TssJ, TssL, and TssM (Durand et al., 2015; Nazarov et al., 2017). Phylogenetic analysis of TssC proteins found that type VI secretion systems cluster into three main groups, where subtypes I and II are proteobacterial, while subtype III is restricted to Bacteroidetes (Russell et al., 2014b). The inner tube is a stack of hexameric Hcp rings capped with a spike complex of a VgrG trimer, further sharpened with a PAAR protein tip; designed for effector and toxin delivery (Leiman et al., 2009; Shneider et al., 2013). T6SSs can directly target both prokaryotic and eukaryotic cells, as well as delivering effector proteins into the extracellular milieu in a contact-independent manner (Pukatzki et al., 2006; Hood et al., 2010; Si et al., 2017b). These effectors display a vast range of activities, including hydrolysis of peptidoglycan of peptidoglycan, nucleic acids, nucleotides, proteins, and lipids; membrane pore formation and metal ion binding, thus conferring a competition advantage to the T6SS-wielding bacterium and promoting its survival (Russell et al., 2014a; Wang et al., 2015; Ahmad et al., 2019). This review examines the relationship between the type VI secretion system and the microbiome in the context of both symbiosis and dysbiosis.
The T6SS Contributes to the Fitness of the Microbiota
The majority of the mammalian microbiome is acquired at birth, with the prevailing species seeded from the mother during delivery and influenced by breastfeeding and environmental exposure (Round et al., 2010). During the first year of life, the composition of the gut microbiome is highly dynamic, in part due to the weaning process, before stabilizing, and remaining consistent through adulthood (Faith et al., 2013; Verster et al., 2017; Al Nabhani et al., 2019). The major constituents of the gut community belong to the phyla Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria, with members of the Bacteroides genus dominating the large intestine (Human Microbiome Project Consortium, 2012). Subtype III of T6SS (hereafter referred as T6SSiii) is restricted to the Bacteroidetes phylum and has been shown to deliver antibacterial effectors resulting in microbial antagonism (Russell et al., 2014b).
Bioinformatic analyses of T6SS loci within the order Bacteroidales has classified them into three distinct “genetic architectures,” designated GA1–3 (Coyne et al., 2016). GA1 and GA2 are found on integrative conjugative elements. Genomic analysis of the co-resident Bacteroides spp. isolated from human gut provided evidence of transfer of these elements between species in situ, implying that T6SS loci are under positive selection in the microbiome (Coyne et al., 2014). GA3 T6SSs are confined to Bacteroides fragilis, an obligate anaerobe, while GA1 and GA2 loci are more widespread within the phylum (Coyne et al., 2016). GA1–3 display distinct repertoires of effector-immunity pairs, possibly driving the incompatibility of these T6SSs within a single niche of an individual (Coyne and Comstock, 2019). One strain of B. fragilis tends to dominate the microbiota of an individual due to strain exclusion as the composition of the community stabilizes (Kostic et al., 2015; Yassour et al., 2016; Verster et al., 2017). Indeed, metagenomic analyses revealed that the abundance of GA3 T6SS loci is higher in infants, suggesting that competition between B. fragilis strains leads to stability of the microbial community in adulthood (Coyte et al., 2015; Verster et al., 2017). These observations should also be considered in light of the weaning process, wherein dietary changes lead to the influx of new bacterial competitors and dietary metabolites required for the host immune ontogeny (Al Nabhani et al., 2019). Co-existence of strains with different T6SSiii “genetic architectures” does arise but solely when bacterial species with overlapping nutritional niches become spatially segregated in the presence of a dense and diverse microbiota (Zitomersky et al., 2011; Hecht et al., 2016).
The use of gnotobiotic mouse models provided the empirical evidence supporting the roles of the T6SS in Bacteroidetes as ecological determinants, wherein T6SS expression and activity have been directly detected in vivo (Russell et al., 2014b; Chatzidaki-Livanis et al., 2016). In vivo competition assays have demonstrated that B. fragilis employs the T6SS to displace competitors from their niche in a contact-dependent manner, with several effector proteins supporting this elimination (Table 1) (Chatzidaki-Livanis et al., 2016; Hecht et al., 2016; Wexler et al., 2016; Ross et al., 2019). Furthermore, in vitro competition assays have found that T6SS-mediated antagonism of Bacteroides spp. targeted a narrow range of species, with most prey strains resistant to intoxication (Chatzidaki-Livanis et al., 2016; Wexler et al., 2016). Thus, the susceptibility to T6SS-dependent antagonism depends as much on the belligerent's identity as on the population distribution across topological niches.
Table 1.
T6SSiii effectors of human symbionts.
| Commensal bacterium | Antibacterial T6SS | Effector locus | Immunity protein(s) | Immunity locus | References |
|---|---|---|---|---|---|
| effector | tag | tag(s) | |||
| Bacteroides dorei DSM 17855 | “GA2_e14” | BACDOR_RS22955 | “GA2_i14” | BACDOR_RS17020 | Ross et al., 2019 |
| Bacteroides fragilis 638R | Bfe1 | BF638R_1988 | Bfi1 | BF638R_1987 | Chatzidaki-Livanis et al., 2016 |
| Bacteroides fragilis 638R | Bfe2 | BF638R_1979 | Bfi2 | BF638R_1978 | Chatzidaki-Livanis et al., 2016 |
| Bacteroides fragilis 638R | – | – | Orphan Bti1 (“GA3_i6”) | BF638R_2042 | Ross et al., 2019 |
| Bacteroides fragilis 638R | – | – | Orphan Bti2a,b (“GA3_i7ab”) | BF638R_2053-4 | Ross et al., 2019 |
| Bacteroides fragilis 638R | – | – | Orphan “GA2_i11” | BF638R_1388 | Ross et al., 2019 |
| Bacteroides fragilis CL03T00C23 | “GA2_e2” | HMPREF1079_RS08215 | “GA2_i2” | HMPREF1079_RS08220 | Ross et al., 2019 |
| Bacteroides fragilis NCTC 9343 | Bte1 (“GA3_e6”) | BF9343_1937 | Bti1 (“GA3_i6”) | BF9343_1936 | Wexler et al., 2016 |
| Bacteroides fragilis NCTC 9343 | Bte2 | BF9343_1928 | Bti2a,b (“GA3_i7ab”) | BF9343_1927-6 | Hecht et al., 2016; Wexler et al., 2016 |
| Bacteroides fragilis NCTC 9343 | – | – | Orphan “GA1_i5” | BF9343_1657 | Ross et al., 2019 |
| Bacteroides fragilis YCH46 | “GA1_e5” | BF2850 | “GA1_i5” | BF2851 | Ross et al., 2019 |
Horizontal gene transfer facilitates the evolution of bacterial species in polymicrobial environments by enabling the positive selection of genes conferring a competitive advantage, a phenomenon also observed for T6SS loci (Unterweger et al., 2014). The existence of “orphan” T6SS immunity genes (conferring resistance to deleterious T6SS effector proteins; bearing no connection to the host immune system) in the absence of cognate effector genes was discovered in Vibrio cholerae isolates, leading to the hypothesis that their acquisition would subsequently protect the bearer against T6SS attacks from non-kin opponents (Kirchberger et al., 2017). The functionality of these orphan immunity genes was elegantly shown by Ross and colleagues in a recent study of members of the microbiome exhibiting extensive arrays called acquired interbacterial defense (AID) clusters (Ross et al., 2019). Here, many members of Bacteroidales were immune to T6SS antagonism by other species and may even possess immunity genes conferring resistance to anti-bacterial effectors associated to strategies beyond the T6SS (Zhang et al., 2012; Ross et al., 2019). However, immunity proteins are not the only way to mitigate the impact of antagonistic effectors. Several studies showed the inability of certain T6SS effectors to intoxicate prey cells lacking the cognate immunity proteins (Altindis et al., 2015; Ringel et al., 2017; Wood et al., 2019), and synergistic effector activities have also been described (LaCourse et al., 2018). Further protection strategies from T6SS-mediated killing, such as upregulation of envelope stress responses and production of extracellular polysaccharides, underscore the complexity of T6SS antagonism (Toska et al., 2018; Hersch et al., 2020).
T6SS-mediated bacterial antagonism targets specific competitors in the gut, helping to dictate niche occupancy. However, when considered in the broader ecological context of the microbiota and symbiosis with the host, the T6SS may also promote the symbiotic relationship with the host by enabling metabolic cooperation (Hooper et al., 2012; Vonaesch et al., 2018). Additionally, the presence of a stable microbiota provides resistance to dysbiosis and outcompetes invading microbial pathogens for nutrients. In terms of direct antibacterial warfare, the T6SS should be considered as a major armament of the microbiota in limiting infection (Kamada et al., 2013; Ducarmon et al., 2019). Indeed, mouse models have shown that the priority benefit of B. fragilis colonization may be protection against infection by enterotoxigenic B. fragilis strains, in a manner that depends on T6SS effector-immunity genotype (Hecht et al., 2016).
Promotion of Immune Homeostasis by the Microbiota: A Potential Role for the T6SS?
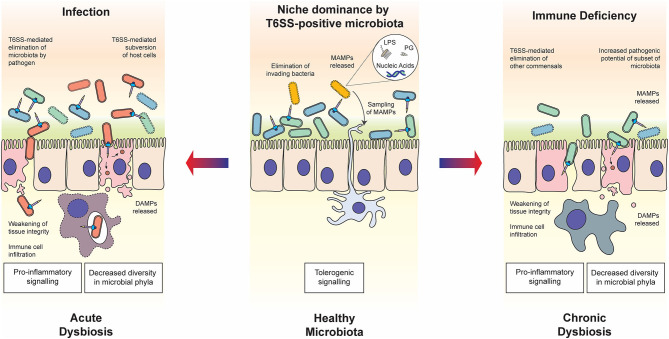
The intestinal microbiota is also crucial for the development of our immune system, as its absence leads to low antibody titer, poor glycosylation of mucosal surfaces, overt TH2 responses and defective development of gut-associated lymphoid tissue in germ-free mice (Smith et al., 2007). The resident microbiota is proposed to train our immune system to actively tolerate the presence of distinct commensals whilst providing robust resistance against invading bacterial pathogens; presenting the intriguing teleological argument that commensal bacteria co-opt the host immune system to defend their niche (Round and Mazmanian, 2009). Evidence now strongly supports the idea of tolerogenic immune responses to commensal flora, rather than specifically ignoring these residents (Round et al., 2010). Tolerance is fostered through the detection of microbe-associated molecular patterns (MAMPs) and microbial metabolites, and extends beyond the local environment of the gut to promote appropriate systemic immune responses (Figure 1) (Clarke et al., 2010; Chu and Mazmanian, 2013). The detection of conserved MAMPs by pattern recognition receptors (PRRs) is one of the foundations of the innate immune system. Innate immune cells, particularly antigen-presenting dendritic cells (DCs) sense their environment in peripheral organs through continuous uptake and sampling of exogenously acquired antigens (Iwasaki and Medzhitov, 2015). Upon microbial encounter, the engagement of PRRs by MAMPs elicits an inflammatory genes program, enhances antigen processing and presentation processes in DCs; all critical for T cell mediated immune responses against pathogens (Medzhitov and Janeway, 1999; Takeuchi and Akira, 2010; Iwasaki and Medzhitov, 2015). MAMPs include lipopolysaccharide (LPS), peptidoglycan, lipoproteins and nucleic acids that trigger MAP kinase and NF-κB signaling leading to pro-inflammatory responses (Fitzgerald and Kagan, 2020). Yet, there is precedent for MAMPs to assist in the development of tolerogenic signals. Mucosal DCs interacting with commensal bacterial components directly or through indirect acquisition of secreted outer membrane vesicles (OMVs) prime host regulatory T cells (Tregs), a subset of T cells promoting tolerance to both food and microbial antigens, thus dampening immune responses to the resident bacterial communities.
Figure 1.
Roles of the T6SS in host-microbiota-pathogen interactions. In healthy steady state conditions (middle panel), commensal bacteria use the T6SS to establish and maintain their niche in the host. The release of MAMPs through T6SS warfare can contribute to the establishment of immune tolerance, enhancing the symbiotic relationship. In the case of host immune deficiency (right panel), for example due to a genetic polymorphism in the host, cross-talk with the microbiota is compromised and the balance within the microbial community may be disrupted, resulting in chronic dysbiosis. The T6SS is likely to play a role in the modulation of competing commensal populations and subsequent decrease in diversity of bacteria phyla, as well as potentially directly manipulating host cells. In the case of infection by pathogenic bacteria wielding a T6SS (left panel), commensal bacteria are eliminated through both direct delivery of antibacterial effectors and indirect mechanisms such as host manipulation and nutrient competition. The state of dysbiosis that follows is acute but may be resolved through elimination of the pathogen by the host immune system. In both states of dysbiosis, the T6SS may play a determining role in eliciting the release of DAMPs, which influences the host immune response.
The homeostasis of the host-microbiota axis is maintained by continuous immune system monitoring (Belkaid and Harrison, 2017). The best characterized example of immune modulation is the production of polysaccharide A (PSA) by B. fragilis, which signals via Toll-like receptor 2 (TLR2) on dendritic cells. This stimulates the differentiation of Tregs, producing an immunosuppressive environment through the secretion of the cytokine IL-10 (Mazmanian et al., 2005; O'Mahony et al., 2008; Round et al., 2011). In IL-10 deficient mice, commensal bacterium Helicobacter hepaticus exhibits colitogenic potential in the presence of gut microbiota, which has been reported to be suppressed by the T6SS of this ε-proteobacterium (Mazmanian et al., 2008; Chow and Mazmanian, 2010; Bartonickova et al., 2013; Jochum and Stecher, 2020). This highlights the interplay of tolerogenic signaling and the T6SS of resident members of the microbiota; however, the mechanistic details of this interaction are yet to be explored.
Tolerogenic immune signaling is also stimulated by commensal metabolites including the short chain fatty acids (SCFAs) acetate, propionate, and butyrate (Parada Venegas et al., 2019); intermediates of vitamin B2 and B9; and amino acid metabolism (Kjer-Nielsen et al., 2012; Venkatesh et al., 2014; Sasabe et al., 2016). Recent work has started to shed light on the numerous benefits that production of SCFAs by commensal bacteria confer to the host. One consequence is the upregulation of oxidative host metabolism by utilization of SCFAs as a carbon source, which bolsters the hypoxic microenvironment at the colonocyte surface, favoring the growth of obligate anaerobes (e.g., Bacteroides spp.) and limiting propagation of facultative aerobes, such as invasive pathogens like Escherichia coli (Litvak et al., 2018; Zhang et al., 2019). In addition, SCFAs act directly via immune cell receptors to modulate T cell subset expansion and macrophage polarization (Schulthess et al., 2019). These compounds commonly promote IL-10 production and suppress inflammation; however, they may also contribute toward effector T cell differentiation, depending on the overall local immunological context (Zhang et al., 2019). On the other hand, microbial metabolites in the intestine may stimulate virulence programs of invading bacteria, with several two-component signal transduction systems in T6SS-positive pathogens having been shown to respond to SCFAs and other metabolites produced by the microbiome (Lawhon et al., 2002; Gonzalez-Chavez et al., 2010; Kohli et al., 2018; Goodman et al., 2020). It is likely that T6SS-mediated turbulent population dynamics occurring during the microbiome development results in variation in the levels of these metabolites. Indeed, bacteria activate diverse antimicrobial programs upon non-kin recognition or danger sensing, including an as-yet uncharacterized diffusible signal from lysed Pseudomonas aeruginosa bacteria that heightens the antibacterial T6SS activity in kin (LeRoux et al., 2015). This antibacterial warfare would further alter levels of microbial products in the local milieu, tipping the ecological balance toward dysbiosis. Moreover, one could hypothesize that bacterial products resulting from the aftermath of T6SS-mediated bacterial antagonism may provide the ligands supporting the development of tolerogenic immune responses. Several lines of evidence from various models lend support to this hypothesis. T6SS-dependent exclusion of Aliivibrio fischeri non-kin strains has been reported during their colonization of the light organs of the Hawaiian bobtail squid Euprymna scolopes (Speare et al., 2018, 2020). The ensuing symbiosis results in morphogenesis of the organs, a process that a combination of A. fischeri LPS and specific monomeric peptidoglycan fragments, issued from cell wall remodeling occurring during bacterial growth and considered as a sign of bacterial viability (referred to as tracheal cytotoxin; TCT), are sufficient to stimulate (Koropatnick et al., 2004). In this case, the peptidoglycan fragments are actively released during A. fischeri growth. In the fruit fly Drosophila melanogaster, recognition of peptidoglycan by the peptidoglycan recognition protein (PGRP) scavenger receptors stimulates the Immune Deficiency (IMD) pathway, similar to that of tumor necrosis factor (TNF) in mammals (Kleino and Silverman, 2014). Alternative isoforms of PGRP can determine bacterial viability: recognition of TCT activates the pathway; whereas recognition of polymeric peptidoglycan fragments (issued from bacterial killing) by a splice variant exerts an inhibitory effect of signal transduction (Neyen et al., 2016). This effectively results in a dampened immune response as reduced pathogen viability could represent a reduced threat. Such interplay also occurs in the intestinal lymphoid tissues, where the generation of IgA-producing B cells is induced following the recognition of gram-negative bacterial peptidoglycan by NOD1 in epithelial cells (Bouskra et al., 2008). Other ligands provide additional cues for microbial viability in host cytosol, such as cyclic dinucleotides sensed by the cGAS-STING and RECON pathways (Moretti and Blander, 2018; Whiteley et al., 2019); and bacterial RNA sensing by TLR8 in the endosome of mammalian epithelial cells (Ugolini et al., 2018).
Equally, it is reasonable to envision T6SS machineries and their effectors as direct inducers of immune tolerance at mucosal sites. In agreement with such possibility, host cells of the innate immune system may forge tolerance by acquiring antigens through OMVs (Shen et al., 2012; Kaparakis-Liaskos and Ferrero, 2015; Chu et al., 2016; Durant et al., 2020). The association of TseF, an iron-acquiring T6SS effector of Pseudomonas aeruginosa, with OMVs may represent an underappreciated role for T6SS effectors in host-microbe interplay (Lin et al., 2017). A better understanding of the activities of T6SS effectors deployed by bacterial species at the interface of mucosal surfaces will illuminate the innate immune sensing and response mechanisms to bacterial molecules released into the host milieu, during homeostasis or under stress conditions.
T6SS Deployment by Bacterial Pathogens: Upsetting the Applecart
By its sheer density, the microbiota offers high resistance to colonization by pathogens. Indeed, pathogens are vastly outnumbered at the start of infection and must compete with the host microbiota for space and nutrients, notwithstanding the contact-dependent and -independent mechanisms of bacterial warfare. Although the T6SS was initially associated with bacterial virulence, the precise role of this apparatus in host infection has not always been clear (Hachani et al., 2016). Recently, studies have highlighted the role of the T6SS in bacterial antagonism during infection, rather than through a direct interaction with host cells. Early evidence for T6SS-mediated competition in vivo emerged from a transposon library screen of Vibrio cholerae strains for impaired colonization of the infant rabbit intestine (Fu et al., 2013). The authors found that tsiV3, encoding the immunity protein to the specialized peptidoglycan hydrolase effector VgrG3, is necessary to alleviate a colonization bottleneck in this model of intestinal infection. Further analysis of T6SS dynamics during V. cholerae colonization found that its role in commensal elimination is largely confined to the jejunum, suggesting that this antibacterial activity may be targeted toward specific microbial residents of this niche (Fu et al., 2018). The T6SS of gastrointestinal pathogens Salmonella enterica serovar Typhimurium and Shigella sonnei are also required for complete virulence, with evidence supporting a role in antagonism of members of the microbiota (Sana et al., 2016; Anderson et al., 2017). Yet, similar to V. cholerae, S. Typhimurium exhibited a limited target range in bacterial competition assays against gram-negative members of the microbiota, again hinting at specific targeting during infection (Sana et al., 2016). Although the abundance of proteobacterial commensals is low in comparison to members of the Bacteroidetes and Firmicutes phyla, they are enriched in many niches, for example Acinetobacter spp. in colonic crypts, and Escherichia and Shigella species in the sigmoid colon (Pédron et al., 2012; James et al., 2020). Due to the clash of nutritional niches between many proteobacterial gut residents and their pathogenic proteobacterial counterparts, T6SS-mediated antagonism is likely to unfold between them. Moreover, metagenomic analyses indicate the presence of species possessing T6SSi components, which are absent from the Bacteroidetes subgroup, thereby supporting the notion of T6SSi-mediated warfare waged by commensal bacteria (Coyne and Comstock, 2019).
The induction of inflammatory host responses is a common mechanism of mass disruption by bacterial competitors, which promotes elimination of the microbial community and dysbiosis (Ackermann et al., 2008). For example, by triggering macrophage pyroptosis, an invasive subpopulation of S. Typhimurium can elicit a large inflammatory response leading to the release of pro-inflammatory cytokines from epithelial cells (Thiennimitr et al., 2012). Although this tissue-invasive S. Typhimurium subpopulation is eliminated by the subsequent infiltration of immune cells, the ensuing inflammatory response (notably the IL-22 signaling axis) reduces iron availability in the lumen. Due to its numerous metal ion acquisition systems, the luminal S. Typhimurium subpopulation is able to outcompete the commensal inhabitants and replicate in the lumen (Behnsen et al., 2014). Similarly, the secretion of cholera toxin by V. cholerae results in iron depletion to favor the pathogen's proliferation at the detriment of the microbiota (Rivera-Chávez and Mekalanos, 2019). The antibacterial activity of the T6SS itself can also stimulate host inflammation. Bacterial lysis mediated by the V. cholerae T6SS in mice mono-colonized with a commensal E. coli strain elicits a host transcriptional response, elevating expression of antimicrobial peptides and NF-κB signaling components (Zhao et al., 2018). NF-κB induction could be recapitulated in vitro using supernatants from T6SS-dependent killing assays, suggesting that MAMPs released from T6SS-mediated bacterial lysis may be the factors supporting the induction of this host response. Furthermore, El Tor pandemic strains of V. cholerae display higher levels of T6SS gene expression than reference clinical isolates, thus underpinning the association of T6SS antibacterial activity with pathology (Zhao et al., 2018). In the TRUC murine model for ulcerative colitis, the presence of a commensal bacterial population promotes spontaneous disease onset in this susceptible host (Garrett et al., 2007). Here, the presence of Proteus mirabilis and Klebsiella pneumoniae in this commensal community correlated with colitogenic potential (Garrett et al., 2010). Both of these species possess T6SSs that display antibacterial activity (Alteri et al., 2013; Hsieh et al., 2019), while this secretion system has also been shown to contribute to the fitness of the pathogens in vivo (Lery et al., 2014; Debnath et al., 2018). One can therefore contemplate a role for this secretion system in the TRUC model whereby T6SS-mediated elimination of commensal bacteria promotes an inflammatory response that cannot be restrained due to the immune genes deficiency of the host, resulting in colitis.
Non-mammalian models also support the notion of T6SS-dependent dysbiosis as a driving force for disease symptoms and pathology. A recent study found that Pseudomonas protegens uses antibacterial effectors to antagonize the gut microbiota of butterfly larvae, enabling tissue invasion and disease onset (Vacheron et al., 2019). Infection of D. melanogaster with V. cholerae results in diarrheal symptoms and gut inflammation (Blow et al., 2005), and the T6SS of the pathogen was found to promote mortality in a manner dependent on the presence of constituents of the microbiota (Fast et al., 2018). The IMD pathway also contributes to this pathology, suggesting that elimination of the fly gut commensal bacteria can be lethal due to exacerbated host inflammatory response (Ryu et al., 2008; Fast et al., 2018). T6SS-mediated depletion of the polymicrobial community impacts tissue repair during fly infection, mirroring the pioneering work establishing the role of the human gut microbiota in tissue homeostasis (Rakoff-Nahoum et al., 2004; Fast et al., 2020).
The competition for nutrients is a key aspect of colonization resistance in the host environment. As discussed above, microbiota niche occupancy is partly dictated by the ability to use specific carbon and nitrogen sources. Around one fifth of the genome of Bacteroides spp. encodes proteins involved in polysaccharide catabolism, conferring great metabolic versatility (Sonnenburg et al., 2005; Schwalm and Groisman, 2017). Besides, the host accentuates the state of nutritional immunity by sequestering metal ions upon infection to limit the replication of pathogens. Recent work by the Shen laboratory and others has revealed a role for the T6SS in nutrient acquisition, whereby the secretion of metal ion-binding proteins facilitates the uptake of zinc, iron, copper or manganese (Wang et al., 2015; Lin et al., 2017; Si et al., 2017a,b; Han et al., 2019). A T6SS-4 mutant of Yersinia pestis exhibited reduced pathogenicity in an orogastric mouse model, indicating the role of this virulence factor in overcoming nutritional immunity during infection (Wang et al., 2015). It is likely that members of the microbiota utilize the T6SS for nutrient acquisition too; however, no T6SS effectors have been described to date. The role of the T6SS of bacterial pathogens in disrupting the steady state of microbiota-host ecosystems is becoming increasingly clear and underscores the importance of the microbiota in colonization resistance alongside the versatility of this secretion system.
Direct Host Cell Contact: T6SS Encounters of the Third Kind
The T6SS versatility extends beyond its prominent antibacterial role in many gram-negative bacteria. As the most evolved member of the contractile injection systems, it delivers effectors into the extracellular milieu or directly into neighboring bacteria and/or eukaryotic targets. Many anti-eukaryotic activities of the T6SS have been described, including the manipulation of biochemical processes governing the physiology of phagocytes and epithelial cells (reviewed in Hachani et al., 2016). Furthermore, several studies have found that the T6SS can target fungal cells, and whereas the human microbiota also harbors fungi such as Candida albicans, these interactions within a host remain unexplored (Haapalainen et al., 2012; Trunk et al., 2018; Storey et al., 2020). A summary of T6SS effector proteins with roles distinct from direct bacterial antagonism are listed in Table 2.
Table 2.
T6SS effectors with roles beyond bacterial antagonism.
| T6SS Effector | Bacterium | Function | References |
|---|---|---|---|
| VgrG1AD | Aeromonas dhakensis | Cytoskeletal disruption | Suarez et al., 2010 |
| TecA | Burkholderia cenocepacia | Inhibition of Rho GTPases | Rosales-Reyes et al., 2012; Aubert et al., 2016 |
| TseZ | Burkholderia thailandensis | Acquisition of Zn2+ | Si et al., 2017a |
| TseM | B. thailandensis | Acquisition of Mn2+ | Si et al., 2017b |
| VgrG5 | Burkholderia pseudomallei; B. thailandensis | Formation of multi-nucleated giant cells | Schwarz et al., 2014; Toesca et al., 2014 |
| EvpP | Edwardsiella piscicida | Inhibition of inflammasome formation | Chen et al., 2017 |
| KatN | Enterohaemorraghic Escherichia coli | Protection against oxidative stress | Wan et al., 2017 |
| OpiA | Francisella tularensis | Phagosomal escape | Eshraghi et al., 2016; Ledvina et al., 2018 |
| Azu | Pseudomonas aeruginosa | Acquisition of Cu2+ | Han et al., 2019 |
| TseF | P. aeruginosa | Acquisition of Fe3+ | Lin et al., 2017 |
| PldA | P. aeruginosa | Internalization into non-phagocytic cells | Jiang et al., 2014 |
| PldB | P. aeruginosa | Internalization into non-phagocytic cells | Jiang et al., 2014 |
| Tle4PA | P. aeruginosa | Disruption of ER homeostasis | Jiang et al., 2016 |
| VgrG2b | P. aeruginosa | Cytoskeletal manipulation | Sana et al., 2012 |
| Tfe1 | Serratia marcescens | Membrane depolarization | Trunk et al., 2018 |
| Tfe2 | S. marcescens | Metabolic dysregulation | Trunk et al., 2018 |
| VgrG1VC | Vibrio cholerae | Cytoskeletal disruption | Pukatzki et al., 2007; Ma et al., 2009 |
| VasX | V. cholerae | Formation of membrane pores | Miyata et al., 2011 |
| YezP | Yersinia pestis | Acquisition of Zn2+ | Wang et al., 2015 |
Once pathogens gain a foothold by ousting the residing microbiota in their desired niche, they must contend with the microbial clearance mechanisms of the host. After phagocytosis by immune cells, phagosomal bacteria are subjected to the oxidative burst, where the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase membrane complex produces superoxide radicals in the vacuole to destroy the engulfed microbe. During oxidative stress, such as after uptake by macrophages, enterohaemorrhagic E. coli (EHEC) secretes the T6SS catalase effector KatN to detoxify the local environment (Wan et al., 2017). Intriguingly, despite being important in the survival of EHEC in macrophages, the absence of KatN did not impact virulence in a streptomycin-treated mouse model. However, the T6SS itself was required for complete virulence suggesting the presence of other host cell-targeted effectors or undescribed compensatory host cell mechanisms. The T6SSs secreting metal binding effectors are upregulated upon oxidative stress, suggesting they likely play a role in defense against reactive oxygen species produced by immune cells (Wang et al., 2015; Lin et al., 2017; Si et al., 2017a,b). Indeed, the zinc-binding effector YezP of Y. pestis is required for intracellular survival in macrophages (Wang et al., 2015).
Burkholderia cenocepacia, an opportunistic pathogen of cystic fibrosis patients, resides primarily in alveolar macrophages where it resists killing (Schwab et al., 2014). The delivery of the T6SS effector TecA into the macrophage cytosol leads to the deamidation of Rho GTPases, which hampers the activity of the NADPH complex (Rosales-Reyes et al., 2012; Aubert et al., 2016). Yet, this inactivation of Rho GTPases is detected by the pyrin inflammasome, leading to caspase-1 activation, pyroptosis and inflammation (Xu et al., 2014). Inflammasomes are vital for enacting cell-autonomous immunity. Thus, they are frequently targeted by invasive bacterial pathogens (Sanchez-Garrido et al., 2020). The NLRC4 and NLRP3 inflammasomes are activated by the type III secretion system of Edwardsiella piscicida after phagocytosis. However, this bacterium is also able to impair the activation of caspase-1 using its T6SS effector EvpP (Chen et al., 2017). The mode of action of this effector remains elusive but appears to prevent the induction of ASC-mediated canonical inflammasome seeding by inhibition of calcium-dependent JNK activation.
The facultative intracellular pathogen Francisella tularensis avoids destruction by macrophages through the action of its T6SS (Nano et al., 2004). Proteomics analysis identified several T6SS effector proteins that are required for escape from the phagosome, and cytosolic replication (Eshraghi et al., 2016). One of these is the phosphatidylinositol 3-kinase (PI3K)-like effector OpiA, which remodels the phospholipid content of the phagosomal membrane to delay its maturation in the endosomal compartment, thereby facilitating pathogen escape prior to lysosomal fusion (Ledvina et al., 2018). The H2-T6SS of Pseudomonas aeruginosa also delivers membrane targeting effector proteins into host cells, namely the phospholipases PldA, PldB, and Tle4 (Jiang et al., 2014, 2016; Wettstadt et al., 2019). While PldA and PldB promote internalization of P. aeruginosa by manipulating the PI3K-Akt signaling axis, Tle4 fragments the endoplasmic reticulum, activating the unfolded protein response and autophagy. However, the benefits of these cellular modifications for the bacterium remain unclear. The T6SS yielded by bacterial pathogens targeting host cells presents a further risk to the microbiota, since the ensuing subversion of host processes affects their ecological niche. Indeed, such indirect impact has been demonstrated in the zebrafish model of cholera, where the actin-crosslinking domain of VgrG1 of V. cholerae stimulates peristalsis, resulting in the collapse of the resident microbial community (Logan et al., 2018). The gradual repopulation by the commensal microbiota may evict the invading pathogen despite the reversal of the numerical advantage; yet the niche must still be conducive for this repopulation to occur.
Conclusions
The extended versatility of the T6SS enriches both the panel of virulence factors of bacterial pathogens, and the mutualism toolkit of symbiotic bacteria. The T6SS plays an underappreciated role in the maintenance of this synergistic steady state in the microbiota. Notwithstanding its original designation as a virulence factor, the T6SS is clearly beneficial to the host in facilitating stable colonization of the microbiota. Further investigation into the genetic architecture of the T6SSiii of Bacteroidales, its target range, and effector-immunity repertoire will provide deeper insight into the ecology of the microbiota. Contact-dependent signaling has been described for CDI toxin delivery into immune prey (Garcia et al., 2016) and analogous processes may also be operated by T6SS effectors targeting both bacteria and eukaryotic cells. Exploring the interactions between the T6SS of commensal bacterial and host cells may illuminate the factors commandeering a homeostatic and balanced tolerogenic signaling; with broader implications in infection, diet, autoimmune and autoinflammatory disorders. In all, we describe the underappreciated roles of the T6SS at the nexus of the microbiota, host and the defense against incoming pathogens; and propose further avenues of investigation to dissect the role of this versatile secretion machine in the establishment and homeostasis of holobionts.
Author Contributions
All authors have intellectually revised this work together and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to all colleagues whose work has not been mentioned in this manuscript due to space limitations.
Footnotes
Funding. TEW is supported by a Harvard Medical School Dean's Innovation Award to Marcia B. Goldberg. EA was supported by MRC (MR/M023230/1) and the Barts Charity (MGU0488) grants. AH was supported by H2020-MSCA-Global Fellowship grant 657766 and NHMRC (GNT1145631).
References
- Ackermann M., Stecher B., Freed N. E., Songhet P., Hardt W.-D., Doebeli M. (2008). Self-destructive cooperation mediated by phenotypic noise. Nature 454, 987–990. 10.1038/nature07067 [DOI] [PubMed] [Google Scholar]
- Ahmad S., Wang B., Walker M. D., Tran H.-K. R., Stogios P. J., Savchenko A., et al. (2019). An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678. 10.1038/s41586-019-1735-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Nabhani Z., Dulauroy S., Marques R., Cousu C., Al Bounny S., Déjardin F., et al. (2019). A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50, 1276–1288.e5. 10.1016/j.immuni.2019.02.014 [DOI] [PubMed] [Google Scholar]
- Alteri C. J., Himpsl S. D., Pickens S. R., Lindner J. R., Zora J. S., Miller J. E., et al. (2013). Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 9:e1003608. 10.1371/journal.ppat.1003608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altindis E., Dong T. G., Catalano C., Mekalanos J. J. (2015). Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair. MBio 6:e00075-15. 10.1128/mBio.00075-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. C., Vonaesch P., Saffarian A., Marteyn B. S., Sansonetti P. J. (2017). Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21, 769–776.e3. 10.1016/j.chom.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Aubert D. F., Xu H., Yang J., Shi X., Gao W., Li L., et al. (2016). A burkholderia type VI effector deamidates Rho GTPases to activate the pyrin inflammasome and trigger inflammation. Cell Host Microbe 19, 664–674. 10.1016/j.chom.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- Bartonickova L., Sterzenbach T., Nell S., Kops F., Schulze J., Venzke A., et al. (2013). Hcp and VgrG1 are secreted components of the Helicobacter hepaticus type VI secretion system and VgrG1 increases the bacterial colitogenic potential. Cell. Microbiol. 15, 992–1011. 10.1111/cmi.12094 [DOI] [PubMed] [Google Scholar]
- Behnsen J., Jellbauer S., Wong C. P., Edwards R. A., George M. D., Ouyang W., et al. (2014). The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40, 262–273. 10.1016/j.immuni.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Harrison O. J. (2017). Homeostatic immunity and the microbiota. Immunity 46, 562–576. 10.1016/j.immuni.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle L. E. H., Bailey C. M., Pallen M. J. (2008). Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8. 10.1016/j.mib.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Blow N. S., Salomon R. N., Garrity K., Reveillaud I., Kopin A., Jackson F. R., et al. (2005). Vibrio cholerae infection of Drosophila melanogaster mimics the human disease cholera. PLoS Pathog. 1:e8. 10.1371/journal.ppat.0010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskra D., Brézillon C., Bérard M., Werts C., Varona R., Boneca I. G., et al. (2008). Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510. 10.1038/nature07450 [DOI] [PubMed] [Google Scholar]
- Chatzidaki-Livanis M., Geva-Zatorsky N., Comstock L. E. (2016). Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. U.S.A. 113, 3627–3632. 10.1073/pnas.1522510113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Yang D., Han F., Tan J., Zhang L., Xiao J., et al. (2017). The bacterial T6SS effector EvpP prevents NLRP3 inflammasome activation by inhibiting the Ca2+-dependent MAPK-Jnk pathway. Cell Host Microbe 21, 47–58. 10.1016/j.chom.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Chow J., Mazmanian S. K. (2010). A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7, 265–276. 10.1016/j.chom.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Khosravi A., Kusumawardhani I. P., Kwon A. H. K., Vasconcelos A. C., Cunha L. D., et al. (2016). Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120. 10.1126/science.aad9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Mazmanian S. K. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675. 10.1038/ni.2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T. B., Davis K. M., Lysenko E. S., Zhou A. Y., Yu Y., Weiser J. N. (2010). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231. 10.1038/nm.2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthurst S. J. (2019). The type VI secretion system: a versatile bacterial weapon. Microbiology 165, 503–515. 10.1099/mic.0.000789 [DOI] [PubMed] [Google Scholar]
- Coyne M. J., Roelofs K. G., Comstock L. E. (2016). Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures , two of which are contained on mobile genetic elements. BMC Genomics 17:58. 10.1186/s12864-016-2377-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. J., Zitomersky N. L., McGuire A. M., Earl A. M., Comstock L. E. (2014). Evidence of extensive DNA transfer between Bacteroidales species within the human gut. MBio 5:e01305-14. 10.1128/mBio.01305-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. J., Comstock L. E. (2019). Type VI secretion systems and the gut microbiota. Microbiol. Spectr. 7, 343–350. 10.1128/microbiolspec.PSIB-0009-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte K. Z., Schluter J., Foster K. R. (2015). The ecology of the microbiome: Networks, competition, and stability. Science 350, 663–666. 10.1126/science.aad2602 [DOI] [PubMed] [Google Scholar]
- Debnath I., Stringer A. M., Smith S. N., Bae E., Mobley H. L. T., Wade J. T., et al. (2018). MrpJ directly regulates Proteus mirabilis virulence factors, including fimbriae and type VI secretion, during urinary tract infection. Infect. Immun. 86:e00388-18. 10.1128/IAI.00388-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducarmon Q. R., Zwittink R. D., Hornung B. V. H., van Schaik W., Young V. B., Kuijper E. J. (2019). Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol. Mol. Biol. Rev. 83:e00007-19. 10.1128/MMBR.00007-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E., Nguyen V. S., Zoued A., Logger L., Péhau-Arnaudet G., Aschtgen M.-S., et al. (2015). Biogenesis and structure of a type VI secretion membrane core complex. Nature 523, 555–560. 10.1038/nature14667 [DOI] [PubMed] [Google Scholar]
- Durant L., Stentz R., Noble A., Brooks J., Gicheva N., Reddi D., et al. (2020). Bacteroides thetaiotaomicron-derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome 8:88 10.1186/s40168-020-00868-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshraghi A., Kim J., Walls A. C., Ledvina H. E., Miller C. N., Ramsey K. M., et al. (2016). Secreted effectors encoded within and outside of the Francisella pathogenicity island promote intramacrophage growth. Cell Host Microbe 20, 573–583. 10.1016/j.chom.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J. J., Guruge J. L., Charbonneau M., Subramanian S., Seedorf H., Goodman A. L., et al. (2013). The long-term stability of the human gut microbiota. Science 34:1237439. 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast D., Kostiuk B., Foley E., Pukatzki S. (2018). Commensal pathogen competition impacts host viability. Proc. Natl. Acad. Sci. U.S.A. 115, 7099–7104. 10.1073/pnas.1802165115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast D., Petkau K., Ferguson M., Shin M., Galenza A., Kostiuk B., et al. (2020). Vibrio cholerae-symbiont interactions inhibit intestinal repair in Drosophila. Cell Rep. 30, 1088–1100.e5. 10.1016/j.celrep.2019.12.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K. A., Kagan J. C. (2020). Toll-like receptors and the control of immunity. Cell 18, 1044–1066. 10.1016/j.cell.2020.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Ho B. T., Mekalanos J. J. (2018). Tracking Vibrio cholerae cell-cell interactions during infection reveals bacterial population dynamics within intestinal microenvironments. Cell Host Microbe 23, 274–281.e2. 10.1016/j.chom.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Waldor M. K., Mekalanos J. J. (2013). Tn-seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host and Microbe 14, 652–663. 10.1016/j.chom.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E. C., Perault A. I., Marlatt S. A., Cotter P. A. (2016). Interbacterial signaling via Burkholderia contact-dependent growth inhibition system proteins. Proc. Natl. Acad. Sci. U.S.A. 113, 8296–8301. 10.1073/pnas.1606323113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bayona L., Comstock L. E. (2018). Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. 10.1126/science.aat2456 [DOI] [PubMed] [Google Scholar]
- Garrett W. S., Gallini C. A., Yatsunenko T., Michaud M., DuBois A., Delaney M. L., et al. (2010). Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300. 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett W. S., Lord G. M., Punit S., Lugo-Villarino G., Mazmanian S. K., Ito S., et al. (2007). Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45. 10.1016/j.cell.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Chavez R., Alvarez A. F., Romeo T., Georgellis D. (2010). The physiological stimulus for the BarA sensor kinase. J. Bacteriol. 192, 2009–2012. 10.1128/JB.01685-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman K. N., Powers M. J., Crofts A. A., Trent M. S., Hendrixson D. R. (2020). Campylobacter jejuni BumSR directs a response to butyrate via sensor phosphatase activity to impact transcription and colonization. Proc. Natl. Acad. Sci. U.S.A. 117, 11715–11726. 10.1073/pnas.1922719117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapalainen M., Mosorin H., Dorati F., Wu R. F., Roine E., Taira S., et al. (2012). Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. tomato DC3000, is required for fitness for competition against bacteria and yeasts. J. Bacteriol. 194, 4810–4822. 10.1128/JB.00611-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani A., Wood T. E., Filloux A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93. 10.1016/j.mib.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Han Y., Wang T., Chen G., Pu Q., Liu Q., Zhang Y., et al. (2019). A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 15:e1008198. 10.1371/journal.ppat.1008198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A. L., Casterline B. W., Earley Z. M., Goo Y. A., Goodlett D. R., Bubeck Wardenburg J. (2016). Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 17, 1281–1291. 10.15252/embr.201642282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch S. J., Watanabe N., Stietz M. S., Manera K., Kamal F., Burkinshaw B., et al. (2020). Envelope stress responses defend against type six secretion system attacks independently of immunity proteins. Nat. Microbiol. 5, 706–714. 10.1038/s41564-020-0672-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R. S., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L. V., Littman D. R., Macpherson A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.-F., Lu Y.-R., Lin T.-L., Lai L.-Y., Wang J.-T. (2019). Klebsiella pneumoniae Type VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 219, 637–647. 10.1093/infdis/jiy534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. (2015). Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353. 10.1038/ni.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K. R., Gomes T., Elmentaite R., Kumar N., Gulliver E. L., King H. W., et al. (2020). Distinct microbial and immune niches of the human colon. Nat. Immunol. 21, 343–353. 10.1038/s41590-020-0602-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Wang X., Wang B., Chen L., Zhao Z., Waterfield N. R., et al. (2016). The Pseudomonas aeruginosa type VI secretion PGAP1-like effector induces host autophagy by activating endoplasmic reticulum stress. Cell Rep. 16, 1502–1509. 10.1016/j.celrep.2016.07.012 [DOI] [PubMed] [Google Scholar]
- Jiang F., Waterfield N. R., Yang J., Yang G., Jin Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15, 600–610. 10.1016/j.chom.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Jochum L., Stecher B. (2020). Label or concept – what is a pathobiont? Trends Microbiol, 28, 789–792. 10.1016/j.tim.2020.04.011 [DOI] [PubMed] [Google Scholar]
- Johansson M. E. V., Larsson J. M. H., Hansson G. C. (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl.), 4659–4665. 10.1073/pnas.1006451107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N., Seo S.-U., Chen G. Y., Núñez G. (2013). Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–35. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- Kaparakis-Liaskos M., Ferrero R. L. (2015). Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 15, 375–387. 10.1038/nri3837 [DOI] [PubMed] [Google Scholar]
- Kirchberger P. C., Unterweger D., Provenzano D., Pukatzki S., Boucher Y. (2017). Sequential displacement of type VI secretion system effector genes leads to evolution of diverse immunity gene arrays in Vibrio cholerae. Sci. Rep. 7:45133. 10.1038/srep45133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Patel O., Corbett A. J., Le Nours J., Meehan B., Liu L., et al. (2012). MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723. 10.1038/nature11605 [DOI] [PubMed] [Google Scholar]
- Kleino A., Silverman N. (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 42, 25–35. 10.1016/j.dci.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli N., Crisp Z., Riordan R., Li M., Alaniz R. C., Jayaraman A. (2018). The microbiota metabolite indole inhibits Salmonella virulence: involvement of the PhoPQ two-component system. PLoS ONE 13:e0190613. 10.1371/journal.pone.0190613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatnick T. A., Engle J. T., Apicella M. A., Stabb E. V., Goldman W. E., McFall-Ngai M. J. (2004). Microbial factor-mediated development in a host-bacterial mutualism. Science 306, 1186–1188. 10.1126/science.1102218 [DOI] [PubMed] [Google Scholar]
- Kostic A. D., Gevers D., Siljander H., Vatanen T., Hyötyläinen T., Hämäläinen A.-M., et al. (2015). The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273. 10.1016/j.chom.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCourse K. D., Peterson S. B., Kulasekara H. D., Radey M. C., Kim J., Mougous J. D. (2018). Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat. Microbiol. 3, 440–446. 10.1038/s41564-018-0113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon S. D., Maurer R., Suyemoto M., Altier C. (2002). Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46, 1451–1464. 10.1046/j.1365-2958.2002.03268.x [DOI] [PubMed] [Google Scholar]
- Ledvina H. E., Kelly K. A., Eshraghi A., Plemel R. L., Peterson S. B., Lee B., et al. (2018). A phosphatidylinositol 3-kinase effector alters phagosomal maturation to promote intracellular growth of Francisella. Cell Host Microbe 24, 285–295.e8. 10.1016/j.chom.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. M., Donaldson G. P., Mikulski Z., Boyajian S., Ley K., Mazmanian S. K. (2013). Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429. 10.1038/nature12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159. 10.1073/pnas.0813360106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoux M., Kirkpatrick R. L., Montauti E. I., Tran B. Q., Peterson S. B., Harding B. N., et al. (2015). Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife 4:e05701. 10.7554/eLife.05701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lery L. M. S., Frangeul L., Tomas A., Passet V., Almeida A. S., Bialek-Davenet S., et al. (2014). Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 12:41 10.1186/1741-7007-12-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Zhang W., Cheng J., Yang X., Zhu K., Wang Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. 10.1038/ncomms14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak Y., Byndloss M. X., Bäumler A. J. (2018). Colonocyte metabolism shapes the gut microbiota. Science 362:eaat9076. 10.1126/science.aat9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan S. L., Thomas J., Yan J., Baker R. P., Shields D. S., Xavier J. B., et al. (2018). The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc. Natl. Acad. Sci. U.S.A. 115, E3779–E3787. 10.1101/226472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A. T., McAuley S. B., Pukatzki S., Mekalanos J. J. (2009). Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5, 234–243. 10.1016/j.chom.2009.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K., Liu C. H., Tzianabos A. O., Kasper D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–18. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L., Kasper D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C. A. (1999). Innate immune induction of the adaptive immune response. Cold Spring Harb. Symp. Quant. Biol. 64, 429–436. 10.1101/sqb.1999.64.429 [DOI] [PubMed] [Google Scholar]
- Miyata S. T., Kitaoka M., Brooks T. M., McAuley S. B., Pukatzki S. (2011). Vibrio cholerae requires the type VI secretion system virulence factor Vasx to kill Dictyostelium discoideum. Infect. Immun. 79, 2941–2949. 10.1128/IAI.01266-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti J., Blander J. M. (2018). Detection of a vita-PAMP STINGs cells into reticulophagy. Autophagy 14, 1102–1104. 10.1080/15548627.2018.1441471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano F. E., Zhang N., Cowley S. C., Klose K. E., Cheung K. K. M., Roberts M. J., et al. (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186, 6430–6436. 10.1128/JB.186.19.6430-6436.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov S., Schneider J. P., Brackmann M., Goldie K. N., Stahlberg H., Basler M. (2017). Cryo-EM reconstruction of type VI secretion system baseplate and sheath distal end. EMBO J. 37:e97103 10.15252/embj.201797103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyen C., Runchel C., Schüpfer F., Meier P., Lemaitre B. (2016). The regulatory isoform rPGRP-LC induces immune resolution via endosomal degradation of receptors. Nat. Immunol. 17, 1150–1158. 10.1038/ni.3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony C., Scully P., O'Mahony D., Murphy S., O'Brien F., Lyons A., et al. (2008). Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-κB activation. PLoS Pathog. 4:e1000112 10.1371/journal.ppat.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ost K. S., Round J. L. (2018). Communication between the microbiota and mammalian immunity. Annu. Rev. Microbiol. 72, 399–422. 10.1146/annurev-micro-090817-062307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada Venegas D., de la Fuente M. K., Landskron G., González M. J., Quera R., Dijkstra G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277 10.3389/fimmu.2019.01486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédron T., Mulet C., Dauga C., Frangeul L., Chervaux C., Grompone G., et al. (2012). A crypt-specific core microbiota resides in the mouse colon. MBio 3:e00116-12. 10.1128/mBio.00116-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513. 10.1073/pnas.0706532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241. 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Ringel P. D., Hu D., Basler M. (2017). The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep. 21, 3927–3940. 10.1016/j.celrep.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Rivera-Chávez F., Mekalanos J. J. (2019). Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 572, 244–248. 10.1038/s41586-019-1453-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Reyes R., Skeldon A. M., Aubert D. F., Valvano M. A. (2012). The type VI secretion system of Burkholderia cenocepacia affects multiple Rho family GTPases disrupting the actin cytoskeleton and the assembly of NADPH oxidase complex in macrophages. Cell Microbiol. 14, 255–273. 10.1111/j.1462-5822.2011.01716.x [DOI] [PubMed] [Google Scholar]
- Ross B. D., Verster A. J., Radey M. C., Schmidtke D. T., Pope C. E., Hoffman L. R., et al. (2019). Human gut bacteria contain acquired interbacterial defence systems. Nature 575, 224–228. 10.1038/s41586-019-1708-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., Lee S. M., Li J., Tran G., Jabri B., Chatila T. A., et al. (2011). The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977. 10.1126/science.1206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., Mazmanian S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., O'Connell R. M., Mazmanian S. K. (2010). Coordination of tolerogenic immune responses by the commensal microbiota. J. Autoimmun. 34, J220–J225. 10.1016/j.jaut.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., Peterson S. B., Mougous J. D. (2014a). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., Wexler A. G., Harding B. N., Whitney J. C., Bohn A. J., Goo Y. A., et al. (2014b). A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236. 10.1016/j.chom.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.-H., Kim S.-H., Lee H.-Y., Bai J. Y., Nam Y.-D., Bae J.-W., et al. (2008). Innate immune homeostasis by the homeobox gene Caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. 10.1126/science.1149357 [DOI] [PubMed] [Google Scholar]
- Sana T. G., Flaugnatti N., Lugo K. A., Lam L. H., Jacobson A., Baylot V., et al. (2016). Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. U.S.A. 113, E5044–E5051. 10.1073/pnas.1608858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana T. G., Hachani A., Bucior I., Soscia C., Garvis S., Termine E., et al. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and fur and modulates internalization in epithelial cells. J. Biol. Chem. 287, 27095–27105. 10.1074/jbc.M112.376368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garrido J., Slater S. L., Clements A., Shenoy A. R., Frankel G. (2020). Vying for the control of inflammasomes: the cytosolic frontier of enteric bacterial pathogen-host interactions. Cell.Microbiol. 22:e13184. 10.1111/cmi.13184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe J., Miyoshi Y., Rakoff-Nahoum S., Zhang T., Mita M., Davis B. M., et al. (2016). Interplay between microbial D-amino acids and host D-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat. Microbiol. 1:16125. 10.1038/nmicrobiol.2016.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess J., Pandey S., Capitani M., Rue-Albrecht K. C., Arnold I., Franchini F., et al. (2019). The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7. 10.1016/j.immuni.2018.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab U., Abdullah L. H., Perlmutt O. S., Albert D., Davis C. W., Arnold R. R., et al. (2014). Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infect. Immun. 82, 4729–4745. 10.1128/IAI.01876-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalm N. D., Groisman E. A. (2017). Navigating the gut buffet: control of polysaccharide utilization in Bacteroides spp. Trends Microbiol. 25, 1005–1015. 10.1016/j.tim.2017.06.009 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Singh P., Robertson J. D., LeRoux M., Skerrett S. J., Goodlett D. R., et al. (2014). VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452. 10.1128/IAI.01368-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Giardino Torchia M. L., Lawson G. W., Karp C. L., Ashwell J. D., Mazmanian S. K. (2012). Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520. 10.1016/j.chom.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider M. M., Buth S. A., Ho B. T., Basler M., Mekalanos J. J., Leiman P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. 10.1038/nature12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M., Wang Y., Zhang B., Zhao C., Kang Y., Bai H., et al. (2017a). The type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition. Cell Rep. 20, 949–959. 10.1016/j.celrep.2017.06.081 [DOI] [PubMed] [Google Scholar]
- Si M., Zhao C., Burkinshaw B. J., Zhang B., Wei D., Wang Y., et al. (2017b). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. U.S.A. 114, E2233–E2242. 10.1073/pnas.1614902114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., McCoy K. D., Macpherson A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69. 10.1016/j.smim.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Sonnenburg J. L., Xu J., Leip D. D., Chen C.-H., Westover B. P., Weatherford J., et al. (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959. 10.1126/science.1109051 [DOI] [PubMed] [Google Scholar]
- Speare L., Cecere A. G., Guckes K. R., Smith S., Wollenberg M. S., Mandel M. J., et al. (2018). Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc. Natl. Acad. Sci. U.S.A. 115, E8528–E8537. 10.1073/pnas.1808302115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speare L., Smith S., Salvato F., Kleiner M., Septer A. N. (2020). Environmental viscosity modulates interbacterial killing during habitat transition. MBio 11, 1–14. 10.1128/mBio.03060-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey D., McNally A., Åstrand M., sa-Pessoa Graca Santos J., Rodriguez-Escudero I., Elmore B., et al. (2020). Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 16:e1007969. 10.1371/journal.ppat.1007969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez G., Sierra J. C., Erova T. E., Sha J., Horneman A. J., Chopra A. K. (2010). A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168. 10.1128/JB.01260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. (2010). Pattern recognition receptors and inflammation. Cell 140, 805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Thiennimitr P., Winter S. E., Bäumler A. J. (2012). Salmonella, the host and its microbiota. Curr. Opin. Microbiol. 15, 108–114. 10.1016/j.mib.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toesca I. J., French C. T., Miller J. F. (2014). The type VI secretion system spike protein VgrG5 mediates membrane fusion during intercellular spread by pseudomallei group Burkholderia species. Infect. Immun. 82, 1436–1444. 10.1128/IAI.01367-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toska J., Ho B. T., Mekalanos J. J. (2018). Exopolysaccharide protects Vibrio cholerae from exogenous attacks by the type 6 secretion system. Proc. Natl. Acad. Sci. U.S.A. 115, 7997–8002. 10.1073/pnas.1808469115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunk K., Peltier J., Liu Y.-C., Dill B. D., Walker L., Gow N. A. R., et al. (2018). The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931. 10.1038/s41564-018-0191-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis R. M., Bäumler A. J. (2020). Gastrointestinal host-pathogen interaction in the age of microbiome research. Curr. Opin. Microbiol. 53, 78–89. 10.1016/j.mib.2020.03.002 [DOI] [PubMed] [Google Scholar]
- Ugolini M., Gerhard J., Burkert S., Jensen K. J., Georg P., Ebner F., et al. (2018). Recognition of microbial viability via TLR8 drives TFH cell differentiation and vaccine responses. Nat. Immunol. 19, 386–396. 10.1038/s41590-018-0068-4 [DOI] [PubMed] [Google Scholar]
- Unterweger D., Miyata S. T., Bachmann V., Brooks T. M., Mullins T., Kostiuk B., et al. (2014). The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat. Commun. 5:3549. 10.1038/ncomms4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron J., Péchy-Tarr M., Brochet S., Heiman C. M., Stojiljkovic M., Maurhofer M., et al. (2019). T6SS contributes to gut microbiome invasion and killing of an herbivorous pest insect by plant-beneficial Pseudomonas protegens. ISME J. 13, 1318–1329. 10.1038/s41396-019-0353-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A. P., et al. (2014). Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity 41, 296–310. 10.1016/j.immuni.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster A. J., Ross B. D., Radey M. C., Bao Y., Goodman A. L., Mougous J. D., et al. (2017). The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe 22, 411–419.e4. 10.1016/j.chom.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonaesch P., Anderson M., Sansonetti P. J. (2018). Pathogens, microbiome and the host: emergence of the ecological Koch's postulates. FEMS Microbiol. Rev. 42, 273–292. 10.1093/femsre/fuy003 [DOI] [PubMed] [Google Scholar]
- Wan B., Zhang Q., Ni J., Li S., Wen D., Li J., et al. (2017). Type VI secretion system contributes to enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS). PLoS Pathog. 13:e1006246. 10.1371/journal.ppat.1006246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Si M., Song Y., Zhu W., Gao F., Wang Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020 10.1371/journal.ppat.1005020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstadt S., Wood T. E., Fecht S., Filloux A. (2019). Delivery of the Pseudomonas aeruginosa phospholipase effectors PldA and PldB in a VgrG- and H2-T6SS-dependent manner. Front. Microbiol. 10:1718. 10.3389/fmicb.2019.01718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler A. G., Bao Y., Whitney J. C., Bobay L.-M., Xavier J. B., Schofield W. B., et al. (2016). Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. U.S.A. 113, 3639–3644. 10.1073/pnas.1525637113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley A. T., Eaglesham J. B., de Oliveira Mann C. C., Morehouse B. R., Lowey B., Nieminen E. A., et al. (2019). Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. 10.1038/s41586-019-0953-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T. E., Howard S. A., Förster A., Nolan L. M., Manoli E., Bullen N. P., et al. (2019). The Pseudomonas aeruginosa T6SS delivers a periplasmic toxin that disrupts bacterial cell morphology. Cell Rep. 29, 187–201.e7. 10.1016/j.celrep.2019.08.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yang J., Gao W., Li L., Li P., Zhang L., et al. (2014). Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513, 237–241. 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- Yassour M., Vatanen T., Siljander H., Hämäläinen A.-M., Härkönen T., Ryhänen S. J., et al. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8:343ra81. 10.1126/scitranslmed.aad0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., de Souza R. F., Anantharaman V., Iyer L. M., Aravind L. (2012). Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7:18. 10.1186/1745-6150-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tang H., Chen P., Xie H., Tao Y. (2019). Demystifying the manipulation of host immunity, metabolism, and extraintestinal tumors by the gut microbiome. Signal Transduct. Target. Ther. 4:41 10.1038/s41392-019-0074-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Caro F., Robins W., Mekalanos J. J. (2018). Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359, 210–213. 10.1126/science.aap8775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomersky N. L., Coyne M. J., Comstock L. E. (2011). Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect. Immun. 79, 2012–2020. 10.1128/IAI.01348-10 [DOI] [PMC free article] [PubMed] [Google Scholar]