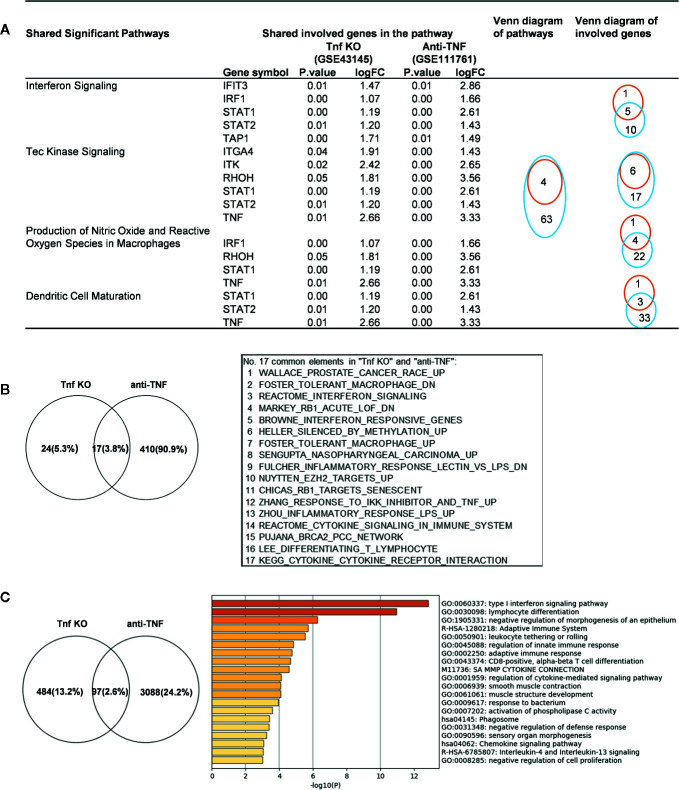
Figure 9.
Up-regulated innatomic genes (IGs) in Tnf KO (GSE43145, Tnf KO Gan mice versus (vs.) Gan mice) and anti-TNF therapy (GSE111761, non-responder vs. responder) microarrays shared pathways. (A) IPA results of the pathways and the involved genes from innatome were compared. Some genes were involved more than two pathways. Orange line represents Tnf KO and blue line represents Anti-TNF therapy. (B) All the significant differential expressed genes (p<0.05, ∣logFC∣>1) of these two microarrays were enrichment analyzed by using GSEA. The result showed a total of 17 pathways were shared by Tnf KO and anti-TNF therapy microarrays. The result showed that except for interferon signaling, several cancer related, inflammatory response and cytokine signalings, were activated in these two microarrays. (C) Venn diagram showed 97 significantly differentially expressed genes were shared by Tnf KO and anti-TNF therapy microarrays. The 97 genes were carried out enrichment analysis by using metascape. The result showed the type I interferon signaling pathway, lymphocyte differentiation, Adaptive Immune System, regulation of innate immune response, response to bacterium etc. were the significantly enriched GO or pathways. Table S10 is IPA results of up- and down-regulated innatomic genes in anti-TNF therapy microarray.