-
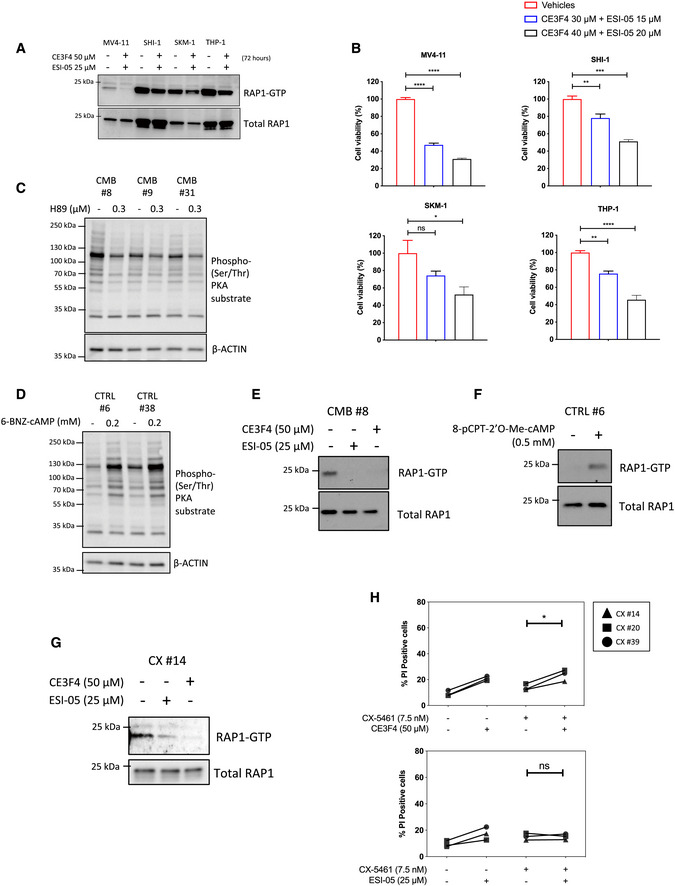
A
Western analysis of the activated, GTP‐bound RAP1 of the indicated human AML cell lines following treatments with the EPAC1 inhibitor CE3F4 and EPAC2 inhibitor ESI‐05 for 72 h (representative of n = 2).
-
B
CellTiterGLO®‐based viability assay of MV4‐11, SHI‐1, SKM‐1, and THP‐1 human AML cells treated as indicated for 72 h. Graphs represent mean ± SEM of n = 3. Data were analyzed by one‐way ANOVA.
-
C, D
Western analysis of PKA activity using the phosphorylated PKA substrate antibody on whole cell extracts isolated from the indicated early passage cell lines treated for 24 h with (C) H89 (n = 3) or (D) 6‐BNZ-cAMP (representatives of n = 3). Actin was used as a loading control.
-
E
Western analysis of the activated, GTP‐bound RAP1 of the early passage CX‐5461 + EV‐resistant (CMB) cell extracts isolated following treatments with indicated EPAC inhibitors for 24 h (representative of n = 2).
-
F
Western analysis of activated, GTP‐bound RAP1 using early passage drug‐naive (CTRL) Eμ‐Myc lymphoma cell extracts isolated following treatments with the selective EPAC activator 8‐pCPT-2‐O-Me-cAMP for 24 h (n = 1).
-
G
Western analysis of the activated, GTP‐bound RAP1 of the early passage CX‐5461-resistant (CX) cell extracts isolated following treatments with indicated EPAC inhibitors for 24 h (n = 1).
-
H
PI exclusion analysis of the CX cells treated with CX‐5461 as indicated in the presence or absence of EPAC1 inhibitor CE3F4 or EPAC2 inhibitor ESI‐05 for 48 h. Data were analyzed by paired one‐way ANOVA. Graphs represent mean ± SEM of n = 3. Black triangle: CX-5461‐resistant (CX) cells clone #14, black square: CX cells clone #20, black circle: CX cells clone #39.
≤ 0.0001.