Figure 3. Antisense oligonucleotides against the GA‐rich region specifically interfere with localization of Rab13 RNA .
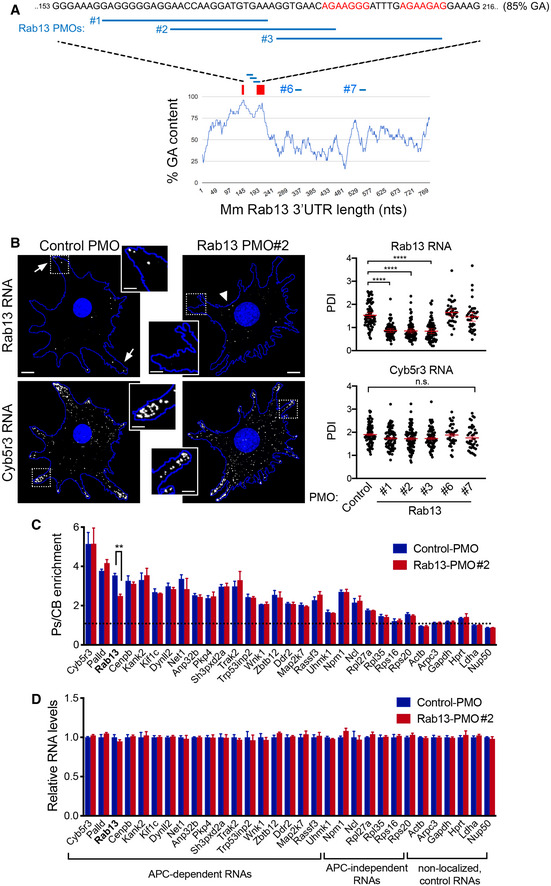
- Schematic showing positions along the mouse Rab13 3′UTR targeted by the indicated PMOs. PMOs #1, 2, and 3 target the RGAAGRR motifs or the adjacent GA‐rich region. PMOs #6 and #7 target the Rab13 3′UTR outside of the GA‐rich region. The control PMO targets an intronic sequence of human β‐globin. Red rectangles and text indicate the location of the GA‐rich motifs.
- FISH images and corresponding PDI measurements of mouse fibroblast cells treated with the indicated PMOs. Cyb5r3 is an APC‐dependent RNA also enriched at protrusions. Arrows: peripheral Rab13 RNA. Arrowheads: perinuclear Rab13 RNA. Boxed regions are magnified in the insets. Note that Rab13 RNA becomes perinuclear in cells treated with PMOs against the GA‐rich region. Scale bars: 10 μm. (4 μm in insets). ****P < 0.0001 by analysis of variance with Dunnett's multiple comparisons test. N = 40–90 cells observed in 3–6 independent experiments. Bars: mean ± 95% CI.
- Protrusion (Ps) and cell body (CB) fractions were isolated from cells treated with control PMO or Rab13‐PMO #2. The indicated RNAs were detected through nanoString analysis to calculate Ps/CB enrichment ratios (n = 3; bars: mean ± s.e.m.). Note that only the distribution of Rab13 RNA is affected. **P = 0.01 by two‐way ANOVA with Bonferroni's multiple comparisons test against the corresponding control.
- Levels of the indicated RNAs were determined using nanoString analysis from control‐ or Rab13 PMO #2-treated cells (n = 4; bars: mean ± s.e.m.). No significant differences were detected by two‐way ANOVA against the corresponding controls.
