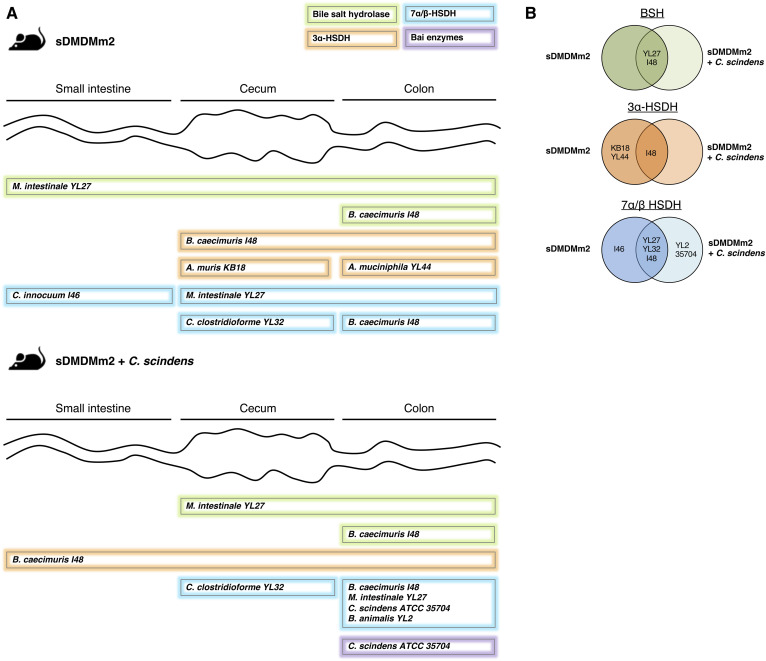
Fig. 3.
Biogeography of specific BA transformation enzymes and the associated microorganisms based on metaproteomic analysis. sDMDMm2 mice (n = 3) and sDMDMm2 mice precolonized 7 days with C. scindens (n = 3) were killed, and intestinal content samples were harvested and processed for metaproteomic analysis. Metaproteomes were mined to find homologous sequences of known BSH, 3α/β-HSDH, 7α/β-HSDH, and 12α-HSDH as well as C. scindens bai genes. A: Scheme illustrating where BSHs, 3α-, 7α-, and 7β-HSDH, and Bai enzymes are expressed in the gut and which bacterial species produce these enzymes. 12α-HSDH and 3β-HSDH were not detected in the metaproteome and therefore are not presented on this figure. Almost all enzymes indicated in panel A were found in the three biological triplicates (supplemental Table S8). The only one that was found in only two mice out of three was the 3α-HSDH from B. caecimuris I48 in the small intestine (supplemental Table S8). B: Venn diagrams summarizing the BA-metabolizing enzymes detected in sDMDMm2 mice and C. scindens-colonized sDMDMm2 mice metaproteomes. For each enzyme detected, the strain producing the enzyme is indicated on the diagram (on the left or right circle if the enzyme was detected in sDMDMm2 mice or C. scindens-colonized sDMDMm2 mice, respectively). If an enzyme was found in both sDMDMm2 and C. scindens colonized-sDMDMm2 mice, the producing strain is indicated in the overlapping part of the two circles.