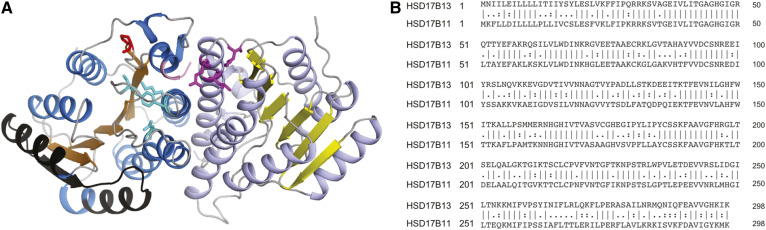
Fig. 3.
Predicted 3D structure of HSD17B13 homodimers using HSD17B11 (67% sequence identity on modeled sequence, PDB entry 1YB1) as the template. A: Structure prediction performed by SWISS-MODEL homology modeling web server. The classical Rossmann-fold structure was observed in the predicted 3D structure of HSD17B13. The β-sheets are colored in orange and yellow, α-helices in marine and light blue, loop regions in gray, amino acids in exon 2 (which are missing in HSD17B13-variant B) in black, catalytic tetrad Asn144-Ser172-Tyr185-Lys189 in cyan, and substrate binding sites Leu199, Glu202, Lys208 in magenta. The Pro260 affected by the P260S mutation is marked in red, and the truncated animo acids affected by the rs72613567-driven G-insertion (only three amino acids are shown in this model) are marked in pink. B: Sequence alignment of human HSD17B13 and HSD17B11 showing overall identity of 191/298 (64.1%) and similarity of 232/298 (77.9%) amino acids.