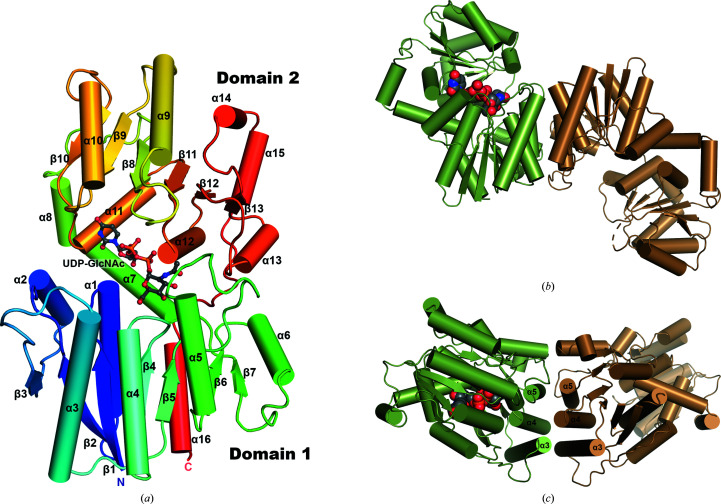
Figure 2.
Structure of NmSacA. (a) Overall structure of the NmSacA monomer bound to the substrate UDP-GlcNAc. The protein is color-coded with a rainbow spectrum from blue at the N-terminus to red at the C-terminus. Secondary-structure elements mentioned in the manuscript are labeled. The UDP-GlcNAc substrate is shown in ball-and-stick representation with gray-colored C atoms. (b) Dimeric structure of one dimer in the crystallographic asymmetric unit of the substrate-bound NmSacA structure. Each dimer only displays UDP-GlcNAc substrate binding (space-filling spheres) in one monomer of each dimer (chain A, green). The substrate-free monomer is in an open conformation (chain B, sand). For clarity, the other dimer (chains C and D) is not shown. (c) Dimer interaction is mediated by three helices (α3, α4 and α5) of each monomer, forming a six-helix bundle at the dimer interface. The UDP-GlcNAc substrate is shown as space-filling spheres with green C atoms (bound to chain A). Similar dimeric interactions are observed in the substrate-free structure.