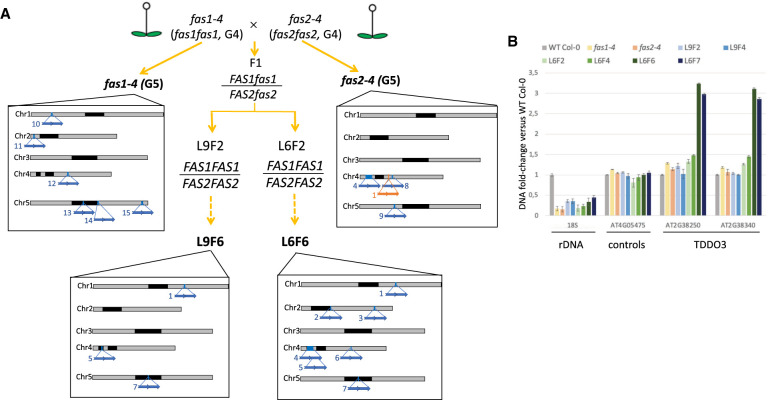
Figure 2.
Identification of TDDO in the parental lines and in L9F6. (A) Identification of TDDO and deletion in the offspring of the parental lines fas1-4 and fas2-4 used to generate the L6 and L9 lines. Lines in bold were sequenced using nanopore technology. Relative distribution and names of each chromosome rearrangement identified are represented by blue (TDDO) and orange (deletion) arrows along chromosomes. Characteristics of each TDDO can be found in Supplemental Table S2. (B) CNV of genes present or not in TDDO3 and of rRNA genes were determined by quantitative PCR. Their relative enrichment was determined in WT Col-0, the parent lines fas1-4 and fas2-4, and in L6 and L9 at several generations. CNVs of rRNA genes were determined using probes amplifying the 18S, the loci AT4G05475 and AT4G16580 that are not duplicated are controls, and the loci AT4TE12140 and AT4G05030 allow the identification of the largest duplication of TDDO4.