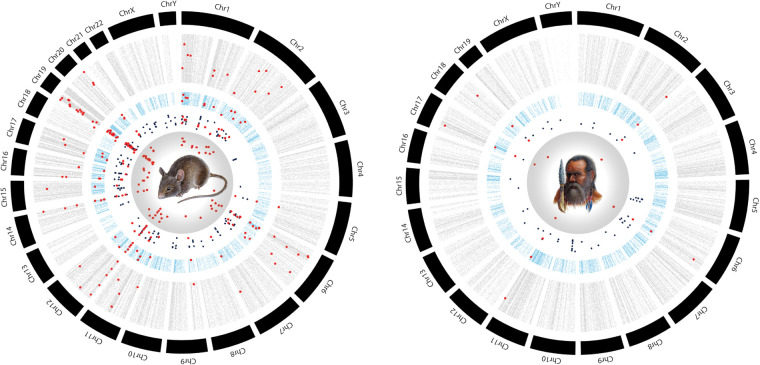
Figure 3.
Consecutive analysis steps for intron loss data sorted by human and mouse chromosomes (for methods, see Gu et al. 2014). The outer black shells show the chromosomal distributions of data points. The gray shells show all screened human introns for intron loss in mouse (left) or mouse introns for intron loss in humans (right). The blue shells represent potential intron losses extracted by 2-n-way under relaxed conditions (allowing perfect presence/absence, presence/presence, and semiperfect cases). The black dots in the white areas represent 2-n-way results from stringent selections of potential intron losses (perfect presence/absence cases). Red dots in the inner circles (containing the species illustrations) represent the manually verified candidate intron loss loci in mouse and human. The manually verified intron losses are highlighted in all the various processing areas.