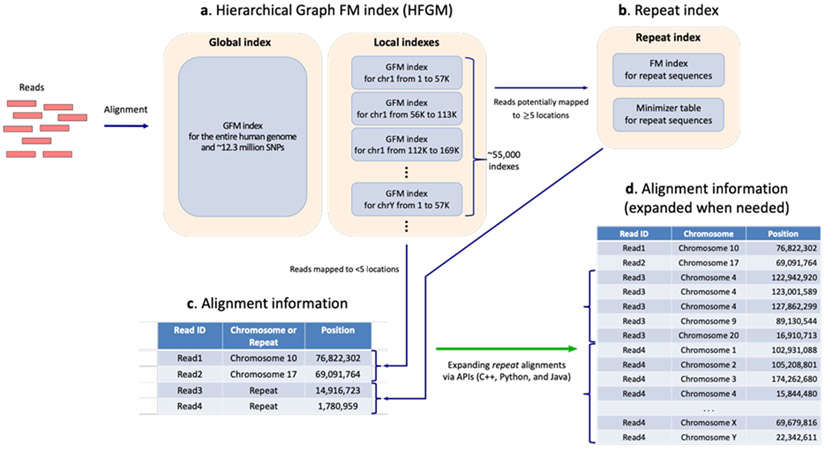
Figure 2.
Overview of HISAT2’s indexes and alignment output
(a) Hierarchical indexing in the hierarchical graph FM index (HGFM). Hierarchical indexing consists of two types of indexes: (1) a global index that represents the entire human genome and (2) 55,172 overlapping local indexes that collectively cover the genome plus all variants. When both are graph FM indexes, a genome plus a large collection of variants can be searched simultaneously. (b) A repeat index represents genomic sequences that are identical. (c) A read matching repeat sequences (e.g., Read3 and Read4) is aligned to just one location (the repeat sequence). (d) The corresponding genomic locations of repeat aligned reads are retrieved via APIs.