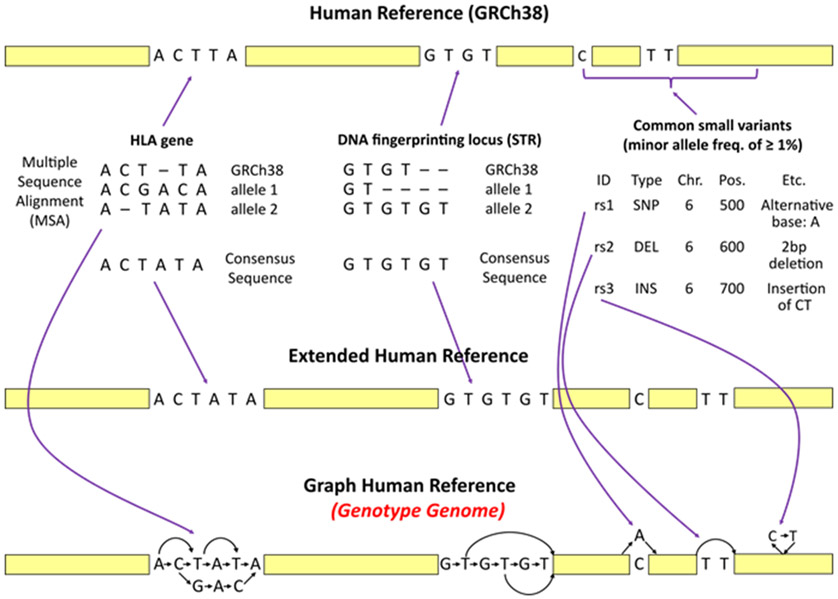
Figure 3.
Construction of the Graph Human Reference, i.e. a Genotype Genome. The figure illustrates how HISAT-genotype extends the human reference genome (GRCh38) by incorporating known genomic variants from several well-studied genes, DNA fingerprinting loci, and common small variants (i.e. variants with minor allele frequencies of ≥1%) from the dbSNP database. In a, the process begins with analyzing information found in the selected databases to construct consensus sequences. The IMGT/HLA database includes over 15,500 allele sequences for 26 HLA genes. A consensus sequence for each HLA gene is constructed based on the most frequent bases that occur in each position of the multiple sequence alignments. The NIST STRBase database contains allele sequences for 13 DNA fingerprinting loci. Because the sequences of the 13 loci are short tandem repeats, HISAT-genotype chooses the longest allele for each locus as a consensus sequence. In b, the human reference is extended by replacing the HLA genes and 13 DNA fingerprinting loci with their consensus sequences. In c, the known genomic variants are then incorporated into the extended references using HISAT2’s graph data structure. Common small variants from dbSNP such as single nucleotide polymorphisms, deletions, and insertions, are also incorporated into the extended reference. In HISAT-genotype this graph reference is called a Genotype genome.