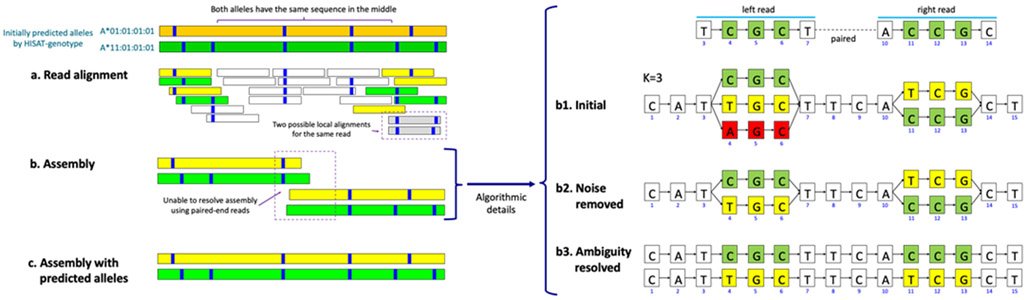
Figure 4.
HISAT-genotype’s assembly of two HLA-A alleles through a guided k-mer assembly graph
The figure shows an abridged example of HISAT-genotype’s assembly output – see Supplementary File 1 for the full assembly output for NA12878. The first two bands are two alleles predicted by HISAT-genotype, in this case A*01:01:01:01 in dark green and A*11:01:01:01 in dark yellow. Each blue stripe indicates where there is a specific genomic variant with respect to the consensus sequence of the HLA-A gene. (a) Shorter bands indicating read alignments whose color is determined according to their degree of compatibility with either of the initially predicted alleles. Reads equally compatible with both alleles are shown in white. Some reads can be locally aligned, i.e. aligned to virtually the same location with just different variants, such as when reads are aligned with or without deletions near their ends, displayed here in gray. (b) Since the two predicted (in fact true/known) alleles share a large common sequence, read pair information is insufficient to fully separate the alleles. HISAT-genotype splits aligned reads into fixed length k-mers. In this simplified case, reads are 5 nucleotides long and k is 3. A pair of reads are aligned at the 3rd location and the 10th location of the graph representation for the HLA gene, respectively. When reads have divergent k-mers, the graph has a corresponding number of branches. One path traversing the graph from left to right constitutes one potential allele sequence. We call this a guided k-mer assembly graph, with guided emphasizing that k-mers are placed according to their aligned locations. The algorithmic details are given in the main text. (c) In addition, HISAT-genotype uses the predicted alleles to enable full-length assembly of both.