Abstract
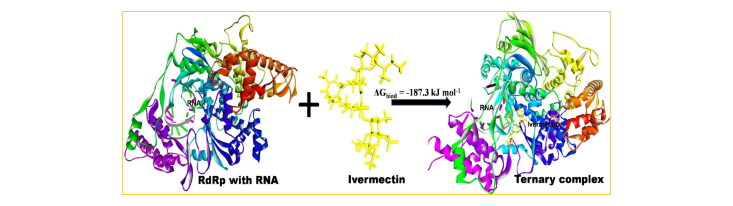
While an FDA approved drug Ivermectin was reported to dramatically reduce the cell line of SARS-CoV-2 by ∼5000 folds within 48 h, the precise mechanism of action and the COVID-19 molecular target involved in interaction with this in-vitro effective drug are unknown yet. Among 12 different COVID-19 targets along with Importin-α studied here, the RNA dependent RNA polymerase (RdRp) with RNA and Helicase NCB site show the strongest affinity to Ivermectin amounting −10.4 kcal/mol and −9.6 kcal/mol, respectively, followed by Importin-α with −9.0 kcal/mol. Molecular dynamics of corresponding protein-drug complexes reveals that the drug bound state of RdRp with RNA has better structural stability than the Helicase NCB site and Importin-α, with MM/PBSA free energy of −187.3 kJ/mol, almost twice that of Helicase (−94.6 kJ/mol) and even lower than that of Importin-α (−156.7 kJ/mol). The selectivity of Ivermectin to RdRp is triggered by a cooperative interaction of RNA-RdRp by ternary complex formation. Identification of the target and its interaction profile with Ivermectin can lead to more powerful drug designs for COVID-19 and experimental exploration.
Keywords: COVID-19, molecular target, ivermectin, antiviral drug, molecular dynamics
Graphical Abstract
Communicated by Ramaswamy H. Sarma
1. Introduction
Covid-19, declared pandemic by WHO (Cucinotta & Vanelli, 2020), is a respiratory disease caused by a novel virus, SARS-CoV-2, which is an enveloped, positive-sense, single-stranded RNA beta-coronavirus. COVID-19 is one of the seven pathogenic members of the coronaviridae family that includes several mild common cold viruses, e.g. hCoV-OC43, HKU, and 229E (Prajapat et al., 2020; Sarma et al., 2020). Looking backward in the recent decade, highly pathogenic human severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) in 2002, and the Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) in 2012, with a fatality rate of 10% and 36%, respectively, have emerged (Chang et al., 2006; Sarma et al., 2020; Zhu et al., 2020). Compared to MERS or SARS, SARS-CoV-2 appears to spread more rapidly, making it difficult to contain that has been a serious concern to the scientific community worldwide now (Gupta et al., 2020; Muralidharan et al., 2020). An increasing number of cases and death from the novel coronavirus worldwide certainly ushers an adverse global impact on health and economics. Considering the fatality and the epidemic nature of this disease, there is a solemn need to find out preventive therapeutics as quickly as possible to curb this virus. However, thus far, no clinically effective drug is approved for the treatment of this virus infection.
SARS-COV-2 has several conserved non-structural and structural proteins (Kong et al., 2020; Wu et al., 2020), which can be the potential targets for the novel or repurposed drug discovery. The non-structural proteins are virus-encoded proteins, but not a part of the viral particle as they express in infected cells. These proteins are typically used for replicating itself by different enzymes and transcription factors. The viral genome is released into host cells as a single-stranded RNA and then translated into viral polyproteins with the help of host protein translation machinery. The polyproteins are further split into a number of non-structural proteins (Nsps) by the main protease (MPro or 3CLpro) and papain-like protease (PLpro). The Nsps play an important role in many processes including the replication of viruses in host cells. Nsp1 and 3 inhibit IFN signaling, blocking the host innate immune response and translation of host’s RNA. Nsp2 with no known function binds to prohibitin proteins; Nsp3 and 5 promote cytokine expression and cleavage of viral polyproteins (Fehr & Perlman, 2015). Similarly, Nsp 12, a conserved protein in coronavirus, is an RNA-dependent RNA polymerase (RdRp) and responsible for coronavirus replication/transcription. Given previous SARS-CoV and MERS-CoV inhibitors’ search considers RdRp as a significant drug target, in the case of SARS-COV-2, Nsp12-RdRp can be most crucial for drug design on which no specific inhibitors have been reported until now (Gao et al., 2020). Together, being conserved and a necessary component for the replication of coronavirus, a multi-functional protein, Nsp13-helicase, is another vital SARS-COV-2 target (Jia et al., 2019), which can be considered further for anti-viral drug discovery provided a very small number of Nsp13 inhibitors reported to date (Cao et al., 2020).
Importin (IMP) is a karyopherin-type protein, which transports protein molecules to the nucleus from the cell cytoplasm. It is of two types, namely Importin α and Importin β. Importin α performs an indispensable role of ferrying proteins from the cytoplasm into the nucleus with a transport carrier, Importin β. Mammalian cells from mouse or human contain either six or seven Importin α subtypes, respectively, each with a tightly regulated expression (Miyamoto et al., 2016). Previously, Ivermectin acted on targets such as IMPα/β1-mediated nuclear import of viral proteins targeting IMPα (Yang et al., 2020). Importin-α targeting compound Ivermectin inhibits by binding to the NLS-binding site of Importin-α, precluding Importin-α/β1 heterodimer formation and subsequently its combination with coronavirus followed by transportation from the cytoplasm to the nucleus through the nuclear pore complex (Atkinson et al., 2018; King et al., 2020; Shechter et al., 2017; Wagstaff et al., 2011; 2012; Yang et al., 2020).
Based on previous evidence of the effectiveness against the earlier SARS, MERS, etc., some FDA approved antiviral drugs remdesivir, lopinavir, ritonavir, and oseltamivir are initially under investigation against COVID-19 (The Scientist dated February 3, 2020. https://www.the-scientist.com/news-opinion/flu-and-anti-hiv-drugs-show-efficacy-againstcoronavirus-67052). Remdesivir, an inhibitor of RNA-dependent RNA polymerase, developed by Gilead to treat Ebola virus infections, is currently in clinical trials for treating COVID-19 (Cao et al., 2020). Recently, a sudden breakthrough in research at Monash University, Australia reporting Ivermectin, an anti-parasitic drug, killing SARS-CoV-2 within 48 h has gained a considerable attention worldwide (Caly et al., 2020). Ivermectin is an FDA-approved drug and has shown in vitro antiviral activity previously against a broad range of viruses, including HIV, Dengue, influenza, and Zika virus (Wagstaff et al., 2012; Yang et al., 2020). A medical team from Bangladesh has claimed that a combination of two widely used drugs Ivermectin and Doxycycline had ‘astounding’ results in curing patients with acute symptoms of SARS-CoV-2 (https://www.trialsitenews.com/bangladesh-medical-college-hospital-physician-see-astounding-results-with-drug-combination-targeting-covid-19/). Now, clinical trial titled ‘Efficacy and Safety of Ivermectin and Doxycycline in Combination or IVE Alone in Patients With COVID-19 Infection’ is going on for the efficacy of said combination of drugs (https://clinicaltrials.gov/ct2/show/NCT04407130). With a lack of further details as to how Ivermectin inhibiting these viruses, the exact mechanism and the target in which Ivermectin interacts with SARS-CoV-2 is yet to be identified (Caly et al., 2020). Such information can proliferate the identification and design of more potent drugs than Ivermectin against SARS-CoV-2 in the near future.
Therefore, the present work explores the interaction between all possible targets of SARS-CoV-2 and Importin-α with Ivermectin to identify the one which is specifically inhibited by the drug providing molecular insights. Molecular docking of Ivermectin with twelve SARS-COV-2’s targets along with Importin-α was carried out, followed by binding mechanism exploration and structural stability analysis using molecular dynamics (MD) simulation through the root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg), and binding free energy of the complexes of Ivermectin with the best targets. The identification of potential target and revelation of the binding mechanism for Ivermectin provide useful information for further exploration in the potential therapeutic discovery against the SARS-CoV-2 pandemic.
2. Materials and methods
2.1. Protein and ligand structure preparation
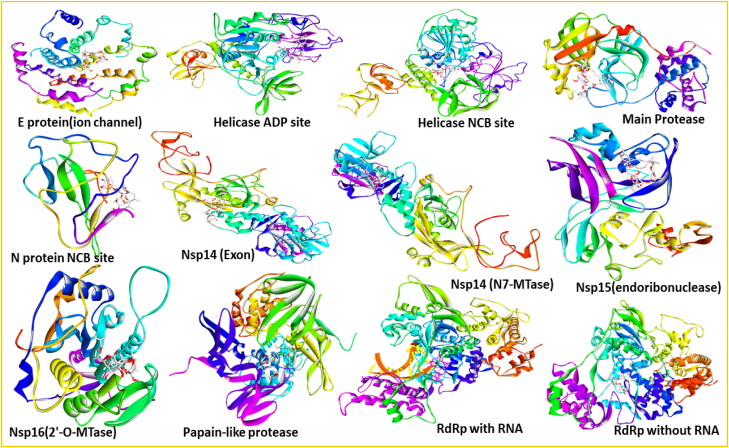
The twelve targets of SARS-CoV-2 used in the study are Main Protease, Papain-like protease, RdRp with RNA, RdRp without RNA, Helicase ADP site, Helicase NCB site, Nsp14(ExoN), Nsp14(N7-MTase), Nsp15(endoribonuclease), Nsp16(2′-O-MTase), N protein NCB site, and E protein(ion channel) (Kong et al., 2020). Apart from them, Importin-α was also used comprising a total of thirteen targets. If available, the structures were retrieved from the RCSB database, otherwise, were modelled using Modeller, see Figure 1. Crystal structures of 6YB7, 6WUU, 7BV2, and 4WV6 for the Main protease, Papain-like protease, RdRp, and Importin-α with high resolutions of 1.25, 1.66, 2.50, and 1.75 Å, respectively, were retrieved from RCSB. The structure of nsp15 in complexation with Uridine-5′-Monophosphate was retrieved from the PDB database with code 6WLC. The structure of nsp16/10 in complexation with 7-methyl-GpppA (GTA), S-Adenosylmethionine (SAM), and 7-methyl-guanosine- 5′-triphosphate (MGP) was retrieved from the PDB database with code 6WVN. The crystal structure of the SARS-CoV-2 nucleocapsid protein with N-terminal RNA-binding domain with PDB id 6M3M was downloaded from RCSB. The modeled structure of E protein was obtained from Zhang’s lab (Zhang et al., 2020). The structure of nsp14 was modeled based on PDB id: 5C8S, the nsp14 structure of SARS-CoV with the sequence identity of 92%. The active site of C-terminal N7-MTase is defined as the SAH-binding site in 5C8S. The model of Helicase was built using crystal structure 6JYT, the helicase structure of SARS-CoV with sequence identity as 98.5%. The structures were validated and energy minimized with Gromacs (Pronk et al., 2013). Downloaded from Drug Bank (Wishart et al., 2018), the SMILES of Ivermectin was converted to the 3 D coordinates using Open Babel (O'Boyle et al., 2011), and next geometry optimization using Gaussian 16 at the level DFT/B3LYP/6-31 + G (d, p).
Figure 1.
The 3 D diagram of Ivermectin binding with twelve different target proteins of SARS-COV-2 in this study.
It is well established from earlier (Atkinson et al., 2018; Shechter et al., 2017; Wagstaff et al., 2011; 2012) as well as recent (King et al., 2020; Yang et al., 2020) experimental studies that Ivermectin binds to Importin-α and not to Importin-β1; inhibition of Importin-α leads to the inhibition of Importin-α/β1 interaction. Thus, in this study, we have used the crystal structure of human Importin-α with the highest resolution and not that of Importin-α/β1 to know the inhibition capability of Ivermectin as the binding site of Ivermectin is present in Importin-α and not in Importin-β1 (Wagstaff et al., 2012; Yang et al., 2020). The interacting residues in the NLS (TAF8)-binding site of Importin-α (PDB id: 4WV6) was visualized and used for the binding of Ivermectin. In 4WV6, the residue number of Importin-α starts from 72 which was, in our study, renumbered from 1. Ivermectin prefers the same site in which chain B (NLS) binds to Importin-α as shown in the superimposed picture of the corresponding protein complexes in Figure S1 (supplementary material). The comparative 2 D plots of Importin-ivermectin interacting residues in the NLS binding site (chain B site) in Figure S2A and S2B (supporting information) clearly indicate no change in the binding-site residues due to renumbering.
Figure 2.
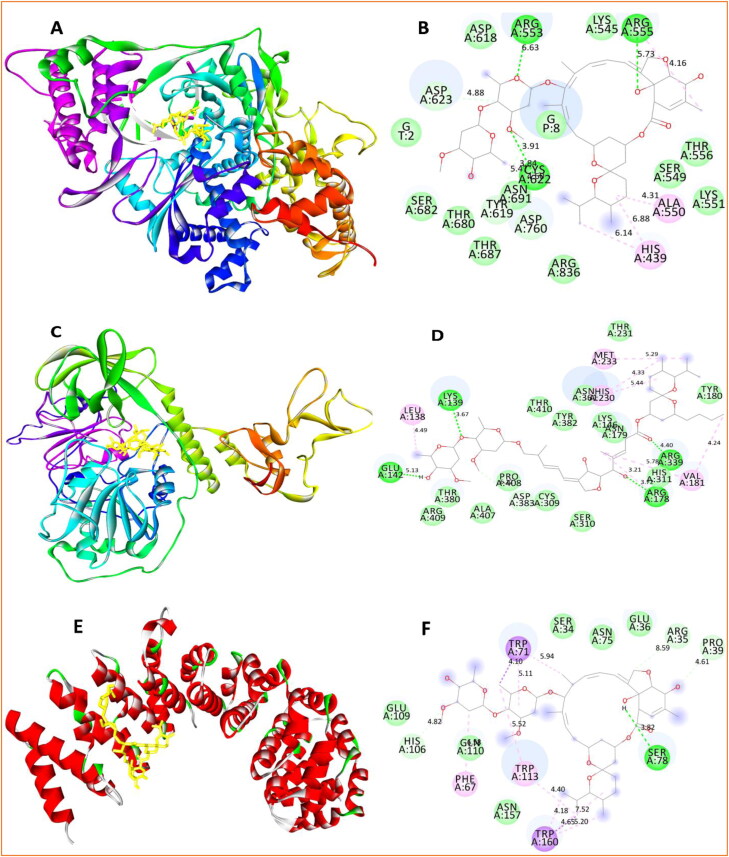
The 3 D and 2 D diagrams depicting, respectively, the target-inhibitor complexes and residues contributed majorly to the interaction between Ivermectin and (a, b) RNA dependent RNA polymerase (RdRp), (c, d) Helicase in the NCB site, and (e, f) Importin-α.
2.2. Molecular docking
Utilizing the COVID-19 docking server which implements AutoDock Vina as a docking engine (Kong et al., 2020), the initial docking of Ivermectin with twelve targets of SARS-CoV-2 was performed, the docking energies and poses of which were later verified manually by AutoDock Vina (Trott & Olson, 2010). The docking of Importin-α with Ivermectin was performed solely using AutoDock Vina. As presented in Table 1, there is an excellent agreement between the docking scores obtained from the COVID-19 docking server and manual docking utilizing AutoDock Vina. The protein targets were prepared by removing water, adding missing hydrogens and Kollman charges to atoms. The addition of hydrogen and assignment of Gasteiger charges and rotatable bonds were performed to Ivermectin. For sequential docking, the Autogrid size was set to particular binding regions of each target with the default grid spacing. Lamarckian Genetic Algorithm (GA 4.2) was used for the docking, which gives the top 10 estimated free energies of binding. The best docking score and pose of complexes were taken, analyzed, and rendered through the Discovery Studio visualizer. The best docked complexes were taken for further molecular dynamics study.
Table 1.
Binding energies (in kcal/mol) of Ivermectin with various targets of SARS-CoV-2 and Importin-α.
| S. No. | SARS-COV-2 targets | Docking server | AutoDock Vina |
|---|---|---|---|
| 1 | E protein (ion channel) | −8.2 | −8.2 |
| 2 | Helicase ADP site | −7.6 | −7.6 |
| 3 | Helicase NCB site | −9.6 | −9.6 |
| 4 | Main Protease | −7.8 | −8.0 |
| 5 | N protein NCB site | −8.3 | −8.3 |
| 6 | Nsp14 (ExoN) | −8.4 | −8.4 |
| 7 | Nsp14 (N7-MTase) | −8.9 | −8.7 |
| 8 | Nsp15 (endoribonuclease) | −6.9 | −6.6 |
| 9 | Nsp16 (2′-O-MTase) | −8.0 | −8.0 |
| 10 | Papain-like protease | −8.5 | −8.4 |
| 11 | RdRp with RNA | −10.4 | −10.4 |
| 12 | RdRp without RNA | −8.7 | −8.8 |
| 13 | Importin-α | −9.0 |
2.3. MD Simulation and MM/PBSA binding free energy
MD simulations were carried out for the RdRp with RNA-Ivermectin, Helicase-Ivermectin, and Importin-α-Ivermectin complexes for a period of 100 ns using GROMACS (Groningen Machine for Chemical Simulations) v5.1 molecular dynamics package (Pronk et al., 2013). The unit cell defined as a cubical box, with a minimal distance of 10 Å from the protein surface to the edges of the box, was solvated using the Simple Point Charge (SPC) water model; the topology of these selected targets and Ivermectin was created by the GROMOS96 53a6 force field (Oostenbrink et al., 2004). Counter-ions were added to make every system electrically neutral at a salt concentration of 0.15 mol/L. Before the MD run, each system was subjected to energy minimization by employing the steepest descent integrator for 5000 steps with force convergence of <1000 kcal mol−1 nm−1.
Thereafter, each protein-ligand complex was equilibrated for 5 ns using canonical (NVT) and isothermal-isobaric (NPT) ensembles. During equilibration, each system was coupled with the Berendsen temperature and Parrinello-Rahman pressure controllers, respectively, to maintain temperature 300 K and pressure 1 bar. The Particle Mesh Ewald (PME) algorithm (Essmann et al., 1995) was employed to deal with the long-range Coulomb interactions with a Fourier grid spacing of 0.12 nm. The short-range van der Waals interactions were given by the Lennard-Jones potential with a cut-off distance of 1 nm. All bond lengths were constrained by the linear constraint solver (LINCS) method (Hess et al., 1997).
Subsequently, 100 ns production run was performed. In principle, the same protocol was applied to all of the systems. A time step of 2 fs was used and the coordinates were saved at every 10 ps during the production run. For the structural analyses of every system, the resultant MD trajectories were analyzed using the built-in modules of GROMACS and visual molecular dynamics (VMD 1.9.1) (Humphrey et al., 1996). The 2 D plots depicting the intrinsic dynamical stabilities captured by the root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg), and principal component analysis (PCA) of the complexes were generated by the Grace 5.1.23 program.
For binding free energy (ΔGbind) from MD trajectory, widely used molecular mechanics/Poisson-Boltzmann surface area (MM/PBSA) was adapted (Kumari et al., 2014; Sen Gupta et al., 2020; Singh et al., 2020). ΔGbind in a solvent medium was calculated as follows:
ΔGbind = Gcomplex - (Gprotein + Gligand), where G comprises the potential energy (EMM) in vacuum and solvation free energy (Gsolv) for each. EMM consists of bonded and non-bonded interactions, whereas Gsolv is the sum of electrostatic and non-polar solvation free energies. The ΔGbind of three complexes was calculated over 10000 frames taken at an equal interval during 100 ns production run.
3. Results and discussion
3.1. Molecular docking and binding mode analysis
Based on docking scores, the interaction of Ivermectin with the twelve targets (as mentioned above; Figure 1) and Importin-α (Figure 2(e)) is ranked as: RNA dependent RNA polymerase (RdRp) with RNA (ΔG = −10.40) > Helicase NCB site (ΔG = −9.60) > Importin-α (ΔG = −9.00) > Nsp14(N7-MTase) (ΔG = −8.90) > RdRp without RNA (ΔG = −8.70) > Papain-like protease (ΔG = −8.50) > Nsp14(ExoN) (ΔG = −8.40) > N protein NCB site (ΔG = −8.30) > E protein (ion channel) (ΔG = −8.20) > Nsp16(2′-O-MTase) (ΔG = −8.00) > Main Protease (ΔG = −7.80) > Helicase ADP site (ΔG = −7.60) > Nsp15(endoribonuclease) (ΔG = −6.90) (Table 1), all ΔGs are in kcal/mol.
Among all targets, RdRp with its cofactors Nsp7_Nsp8 and RNA has the best docking score with Ivermectin, −10.40 kcal/mol, followed by Helicase in NCB site (-9.60 kcal/mol) and Importin-α (ΔG = −9.00 kcal/mol) (Table 1). ΔG below or equal to −9.0 kcal/mol demonstrates a significant interaction based on which three targets, namely RdRp with RNA, Importin-α, and Helicase in NCB site are shortlisted for further investigation. The amino acid residues of these targets interacting with Ivermectin are provided in Table S1 (supplementary material). The residues of the best scoring targets revealed in Table S2 (supplementary material), involve in hydrogen bond (h-bond) formation and π interactions. In the case of RdRp, ARG553, ARG555, and CYS622 participate in h-bond formation, whereas HIS439 and ALA550 take part in π interactions.
Besides, according to Figure 2a,b for the RdRp-Ivermectin complex, RNA molecule interacts with Ivermectin via the formation of a ternary complex, providing more stability.
The energy contribution of RNA to the RdRp-Ivermectin complex is −1.70 kcal/mol. In Figure 2b, the residues predominantly interacting via van der Walls interaction are LYS545, THR556, SER549, LYS551, ASP618, ASP623, SER682, THR680, TYR619, ASN691, ASP760, THR687, and ARG836. On the other hand, residues involved in h-bond formation in the Helicase-Ivermectin complex are LYS139, GLU142, ARG178, and ARG339 (Figure 2c,d; Table S1, supplementary material). Π-interacting residues are LEU138, VAL181, HIS230, and MET233. Similar to the RdRp-Ivermectin complex, residues majorly involved in van der Waals interaction in the Helicase-Ivermectin complex are TYR180, THR231, ASN179, ASN361, THR410, TYR382, LYS146, HIS311, SER310, CYS309, PRO408, ALA407, THR380, and ARG409. In the case of the third complex, Importin-α-Ivermectin (Figure 2(e, f)), there is only one h-bond forming residue which is SER78. Π-interacting residues are PHE67, TRP113, TRP71, and TRP160. Similar to other complexes, the van der Waals interaction is majorly contributing to the Importin-α-Ivermectin complex; the residues involved are SER34, ARG35, GLU36, PRO39, ASN75, HIS106, GLU109, GLN110 and ASN157. Thus, from the interaction point of view, it is found to have four, three, and one h-bond in the Helicase-Ivermectin, RdRp-Ivermectin, and Importin-α-Ivermectin complexes, respectively. In addition to a greater number of h-bonds, the lower binding energy or more stability of RdRp-Ivermectin compared to Helicase-Ivermectin and Importin-α-Ivermectin stems from a cooperative interaction of −1.70 kcal/mol due to the ternary complex formation of RNA with RdRp-Ivermectin.
3.2. MD Trajectory analysis
Molecular Dynamics (MD) simulations have great significance to analyze the internal motions, conformational changes, stability, etc., of protein-ligand complexes ( Sen Gupta et al., 2020; Singh et al., 2020 ). Utilizing the MD trajectories generated, RMSD, RMSF, and Rg were computed and the results are discussed below.
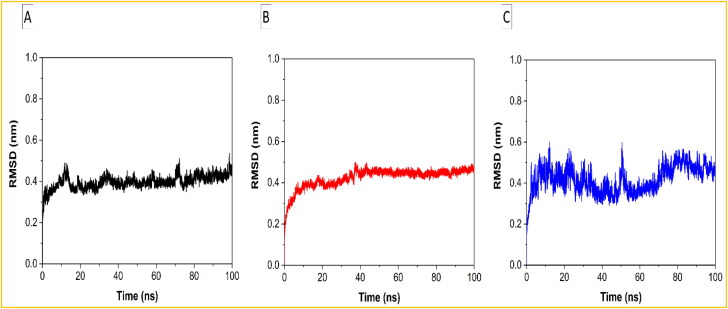
Apart from allowing to assess the equilibration, quality of the run and convergence of MD trajectories, RMSD is useful to investigate the stability of a protein in complex ( Sen Gupta et al., 2020; Singh et al., 2020 ). A larger RMSD value is indicative of the lower stability of a protein complex. Here, the RMSD of the Helicase-Ivermectin, RdRp-Ivermectin, and Importin-α-Ivermectin complexes with respect to the Cα atom was calculated against the MD simulation time and is shown in Figures 3a, b, and c, respectively. In the case of the Helicase-Ivermectin complex, the average value of RMSD is around 0.43 nm, with fluctuations at around 10-15 ns, 33 ns, and 70-75 ns, suggesting a loss of stability in these regions (Figure 3(a)). In the case of the RdRp-Ivermectin complex, the average value of RMSD is around 0.45 nm and almost no major fluctuation has been seen during 100 ns simulation period, suggesting greater stability of the complex throughout the whole dynamics (Figure 3(b)). In the case of Importin-α-Ivermectin, the average value of RMSD is around 0.47 nm, with larger oscillations (Figure 3(c)) while compared to the other two complexes, suggesting its least stability among the three complexes.
Figure 3.
Plots of RMSD as a function of time for the (a) Helicase NCB site-Ivermectin, (b) RdRp-RNA-Ivermectin, and (c) Importin-α-Ivermectin complexes.
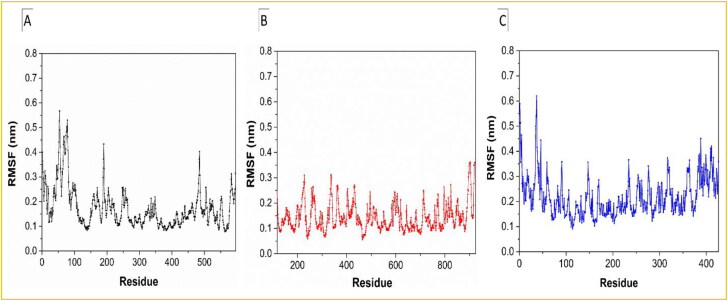
RMSF is a dynamical parameter that measures the average main chain flexibility at each residue position. RMSF measured with respect to backbone atoms of each amino acid residue for all of the three complexes is presented in Figure 4a–c. In the case of the Helicase-Ivermectin complex, the average value of RMSF is around 2.7 nm and higher spikes are noticed at the residues 50 to 90, 190, and 490 (Figure 4(a)). But, in the case of the RdRp-Ivermectin complex, smaller spikes rather can be seen (Figure 4(b)), which corroborates with the RMSD analysis, indicating the more stability of the complex. On the other hand, similar to Helicase-Ivermectin, more frequent fluctuations can be seen in the case of the Importin-α-Ivermectin complex indicating less stability.
Figure 4.
Plots of RMSF as a function of amino acid residues for the (a) Helicase NCB site-Ivermectin, (b) RdRp-RNA-Ivermectin, and (c) Importin-α-Ivermectin complexes.
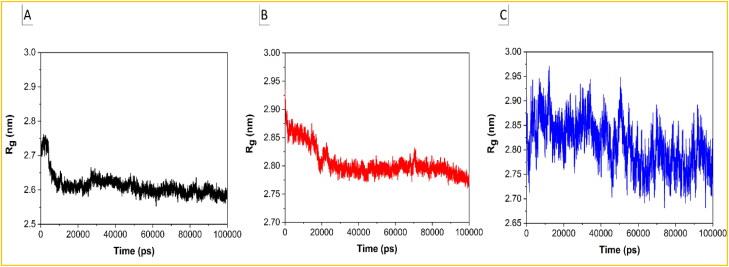
The radius of gyration (Rg) describes the level of compaction of protein. It is defined as the mass-weighted root-mean-square distance for a collection of atoms from their common center of mass. Hence, the trajectory analysis for Rg provides the overall dimension of protein. In the case of the Helicase-Ivermectin complex, the average value of Rg is 2.65 nm and there is a large fall at around 4.9 ns (Figure 5(a)). In the case of the RdRp-Ivermectin complex, the average value of Rg is around 2.8 nm and a small downward jump can be seen at around 19 ns before getting stabilized (Figure 5(b)).
Figure 5.
Plots of Rg as a function of simulation time for the (a) Helicase NCB site-Ivermectin, (b) RdRp-RNA-Ivermectin, and (c) Importin-α-Ivermectin complexes.
The larger average value of Rg for the RdRp-Ivermectin complex may be attributed to its bigger size consisting of a greater number of residues than the Helicase-Ivermectin complex. Despite having a greater number of residues, the fluctuation is lesser than that of the Helicase-Ivermectin complex. In accordant with RMSD and RMSF, the larger average value of Rg of 2.87 nm and fluctuations of the Importin-α-Ivermectin complex (Figure 5(c)) imply its least stability among all the complexes. Altogether, the analysis of RMSD, RMSF, and Rg suggests better stability of the RdRp-Ivermectin than Helicase-Ivermectin or Importin-α-Ivermectin complexes, validating the preliminary docking results. Thus, RdRp with RNA is the most plausible target for Ivermectin.
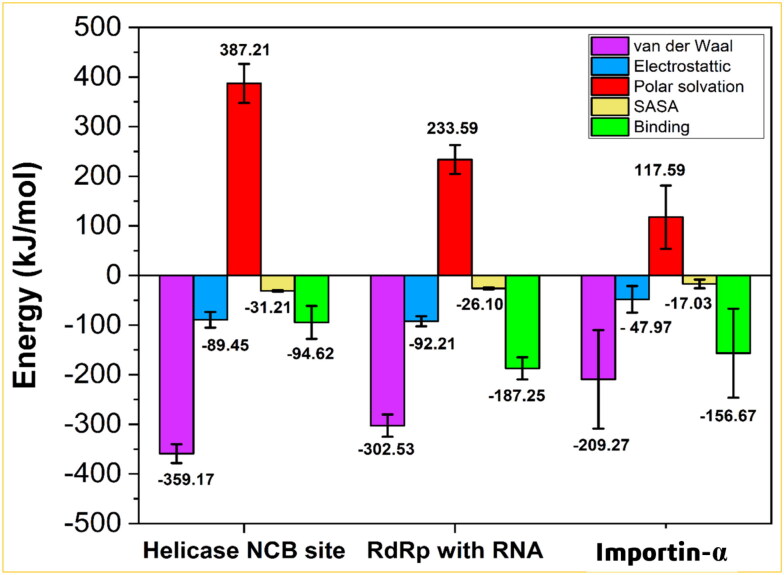
The binding free energy (ΔGbind) has been computed with MM/PBSA (Kumari et al., 2014), which is considered as a very efficient and reliable approach to study crucial molecular recognition processes (Kumari et al., 2014; Open Source Drug Discovery Consortium, 2014;Sen Gupta et al., 2020; Singh et al., 2020). Figure 6 and Table S2 (supplementary material) reveal that the RdRp-Ivermectin complex has ΔGbind (-187.3 kJ mol−1) twice as high as the Helicase-Ivermectin complex (-94.6 kJ mol−1), even better than the Importin-α-Ivermectin complex (-156.7 kJ mol−1). This striking difference in ΔGbind reflects a keen affinity of Ivermectin to RdRp-RNA than Helicase in NCB site or Importin-α. In the decomposition of ΔGbind into different interactions presented in Figure 6 and Table S2 (supplementary material), it is evident that the polar solvation energy is positive in all of the three complexes, thereby opposing the binding and increasing the total binding free energy of the complexes.
Figure 6.
The van der Waals (ΔEvdW), electrostatic (ΔEelec), polar solvation, solvent accessible surface area (SASA) energies and the binding free-energy (ΔGbind) in kJ/mol are represented as a bar plot with error bar for all of the three complexes.
Among other interactions, the van der Waals (ΔEvdW) and solvent-accessible surface area (SASA) energies are the most and the least favourable contributions to the negative binding free energy of all of the complexes (Figure 6). As obvious, a smaller penalty of polar solvation energy and more negative electrostatic energy give rise to the most negative binding free energy for the RdRp-Ivermectin complex among all, confirming its more stability over the other two complexes.
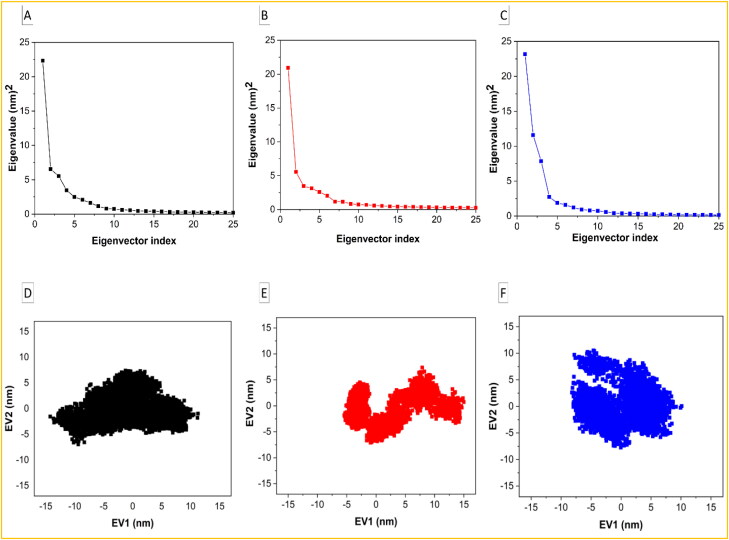
The trajectories of MD simulations were used for principal component analysis (PCA) to identify the conformational motions relevant to protein functions. Eigenvalues were mostly used to calculate the conformational changes due to the movement of atoms (Khan et al., 2016), and were generated by diagonalizing the covariance matrix of the Cα atomic fluctuations against the equivalent eigenvectors’ (EVs) indices. The first 20 modes were taken into consideration in the analysis of the essential subspace as they cover >95% variance of the protein where an exponentially decaying curve of eigenvalues is obtained against the EVs (Figures 7(a)–(c)). In this study, PC1 and PC2 that dominate the protein conformational fluctuations were also used for the analysis of the Helicase-Ivermectin (Figure 7(a, d)), RdRp-Ivermectin (Figure 7(b, e)), and Importin-α-Ivermectin (Figure 7(c, f)) complexes.
Figure 7.
Principal component analysis (PCA) of the Helicase-Ivermectin (Black color), RdRp-Ivermectin (Red color), and Importin-α-Ivermectin (Blue color) complexes. (a, b, and c) - the plot of eigen values versus the corresponding eigen vector indices coming from the Cα covariance matrix during MD simulations and (d, e, and f) - the 2 D projection plots of the first two principal eigenvectors.
Taken the first two PCs into consideration, simulation results revealed the subspace dimension of the complexes in which RdRp-Ivermectin is the thermodynamically most stable one because of the lowest subspace dimension (Figure 7(d)–(f)). This is also reflected in their 2 D projection plots of trajectories (Figure 7(a)–(c)), based on the trace values of their covariance matrixes.
The overall analysis manifests that the RdRp-Ivermectin and Helicase-Ivermectin complexes have retained their stability as reflected from their least conformational changes due to decreasing collective motions.
Despite identifying suitable targets such as Helicase, RNA dependent RNA polymerase (RdRp), and Importin-α, the reported key residues and RNA involved in the interaction with Ivermectin here could be used for further development and improvement of drugs for the mentioned targets. Previously, for other RNA viruses, Ivermectin acted on targets such as Importin-α-mediated nuclear import of viral proteins (Yang et al., 2020) and Helicase (Mastrangelo et al., 2012). Inhibition of the Importin-α/β1 nuclear import pathway by targeting Importin-α has been reported to be the mode of action of Ivermectin (Atkinson et al., 2018; King et al., 2020; Shechter et al., 2017; Wagstaff et al., 2011; 2012; Yang et al., 2020). In this study, the identification of RdRp as a potential target for Ivermectin for SARS-COV-2 would certainly bring more opportunities to resist this virus. Further improvement of antiviral drugs could be possible by functional group modification according to the hydrophobicity and hydrophilicity in the active site region containing the interacting residues, ligand, and RNA as shown in Figure S3 (supporting information). The modification of hydrophilic groups of the ligand in the hydrophobic region and vice-versa would enhance interactions, thus the inhibitory power of the drug. Besides, the computational protocol applied here and the outcome would trigger target-driven drug discovery for SARS-COV-2 which seems to be the need of the hour and should promote studies for other pathogens.
4. Conclusion
Ivermectin, an FDA-approved drug, has shown in vitro antiviral activity against SARS-COV-2. But the exact mechanism and the specific target which Ivermectin inhibits were not known. In this study, analyses of molecular docking revealed that out of twelve molecular targets of SARS-COV-2 and Importin-α, three targets (RdRp with RNA, Helicase in NCP site, and Importin-α) have better binding energies with Ivermectin, lesser than or equal to −9.0 kcal/mol. Comparing among the three complexes of Ivermectin with the shortlisted targets, the RMSD, RMSF, Rg, and the binding free energy from MD demonstrate a greater stability of the RdRp-Ivermectin complex than that of Helicase or Importin-α. The presence of RNA augments the interaction between RdRp and Ivermectin. Thus, RdRp is identified as the most probable target for the in-vitro effective FDA approved drug Ivermectin, which can guide the experiment as well as proliferate the design of more potential therapeutics against SARS-CoV-2.
Supplementary Material
Acknowledgements
P.S.S.G also sincerely acknowledges IISER Berhampur for providing him the Institute Postdoc Fellowship to carry out this work.
Funding Statement
The authors acknowledge IISER Berhampur for computational support.
Disclosure statement
The authors report no conflicts of interest.
References
- Atkinson, S. C., Audsley, M. D., Lieu, K. G., Marsh, G. A., Thomas, D. R., Heaton, S. M., Paxman, J. J., Wagstaff, K. M., Buckle, A. M., Moseley, G. W., Jans, D. A., & Borg, N. A. (2018). Recognition by host nuclear transport proteins drives disorder-to-order transition in Hendra virus V. Scientific Reports, 8(1), 1–7. doi: 10.1038/s41598-017-18742-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., & Wagstaff, K. M. (2020). The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178, 104787. Apr 3: 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y.-C.,, Deng, Q.-X., &, Dai, S.-X. (2020. May- Jun). Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel Medicine and Infectious Disease, 35, 101647. .10.1016/j.tmaid.2020.101647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C., Sue, S.-C., Yu, T., Hsieh, C.-M., Tsai, C.-K., Chiang, Y.-C., Lee, S., Hsiao, H., Wu, W.-J., Chang, W.-L., Lin, C.-H., & Huang, T. (2006). Modular organization of SARS coronavirus nucleocapsid protein. Journal of Biomedical Science, 13(1), 59–72. 10.1007/s11373-005-9035-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta, D., & Vanelli, M. (2020). WHO declares COVID-19 a pandemic. Acta Bio-Medica : Atenei Parmensis, 91(1), 157–160. 10.23750/abm.v91i1.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses. Methods in Molecular Biology (Clifton, N.J.).), 1282, 1–23. 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Yan L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., & Ge, J. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science, 368(6492), 779–82. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 38, 1–7. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, U., Blairy, S., & Kleck, R. E. (1997). The intensity of emotional facial expressions and decoding accuracy. Journal of Nonverbal Behavior, 21(4), 241–257. Dec 1 DOI: 10.1023/A:1024952730333 [DOI] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. DOI: 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jia, Z., Yan, L., Ren, Z., Wu, L., Wang, J., Guo, J., Zheng, L., Ming, Z., Zhang, L., Lou, Z., & Rao, Z. (2019). Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Research, 47(12), 6538–6550. Jul 9 10.1093/nar/gkz409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, Y., Garg, M., Gui, Q., Schadt, M., Gaikwad, A., Han, D., Yamamoto, N. A. D., Hart, P., Welte, R., Wilson, W., Czarnecki, S., Poliks, M., Jin, Z., Ghose, K., Egitto, F., Turner, J., & Arias, A. C. (2016). Flexible hybrid electronics: direct interfacing of soft and hard electronics for wearable health monitoring. Advanced Functional Materials, 26(47), 8764–8775. 10.1002/adfm.201603763 [DOI] [Google Scholar]
- King, C. R., Tessier, T. M., Dodge, M. J., Weinberg, J. B., & Mymryk, J. S. (2020). Inhibition of human adenovirus replication by the importin α/β1 nuclear import inhibitor ivermectin. Journal of Virology, 94(18), eD07. https://doi.org/ 10.1128/JVI.00710-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, R., Yang, G., Xue, R., Liu, M., Wang, F., Hu, J., Guo, X., & Chang, S. (2020). COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics, btaa645. 10.1093/bioinformatics/btaa645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., & Lynn, A., Open Source Drug Discovery Consortium (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations . Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Mastrangelo, E., Pezzullo, M., De Burghgraeve, T., Kaptein, S., Pastorino, B., Dallmeier, K., de Lamballerie, X., Neyts, J., Hanson, A. M., Frick, D. N., Bolognesi, M., & Milani, M. (2012). Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: New prospects for an old drug. The Journal of Antimicrobial Chemotherapy, 67(8), 1884–1894. Aug 1 10.1093/jac/dks147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, Y., Yamada, K., & Yoneda, Y. (2016). Importin α: A key molecule in nuclear transport and non-transport functions. Journal of Biochemistry, 160(2), 69–75. 10.1093/jb/mvw036 [DOI] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 38, 1–7. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox . Journal of Cheminformatics, 3(1), 33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenbrink, C., Villa, A., Mark, A. E., & van Gunsteren, W. F. (2004). A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem, 25(13), 1656–1676. 10.1002/jcc.20090 [DOI] [PubMed] [Google Scholar]
- Prajapat, M., Sarma, P., Shekhar, N., Avti, P., Sinha, S., Kaur, H., Kumar, S., Bhattacharyya, A., Kumar, H., Bansal, S., & Medhi, B. (2020). Drug targets for corona virus: A systematic review. Indian Journal of Pharmacology, 52(1), 56–65. 10.4103/ijp.IJP_115_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk, S., Páll, S., Schulz, R., Larsson, P., Bjelkmar, P., Apostolov, R., Shirts, M. R., Smith, J. C., Kasson, P. M., van der Spoel, D., Hess, B., & Lindahl, E. (2013). GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics (Oxford, England)), 29(7), 845–854. 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma, P., Sekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Prakash, A., Dhibar, D. P., & Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 38, 1. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Gupta, P. S., Islam, R. N., Banerjee, S., Nayek, A., Rana, M. K., & Bandyopadhyay, A. K. (2020). Screening and molecular characterization of lethal mutations of human homogentisate 1, 2 dioxigenase. Journal of Biomolecular Structure and Dynamics, 38, 1. 10.1080/07391102.2020.1736158 [DOI] [PubMed] [Google Scholar]
- Shechter, S., Thomas, D. R., Lundberg, L., Pinkham, C., Lin, S. C., Wagstaff, K. M., Debono, A., Kehn-Hall, K., & Jans, D. A. (2017). Novel inhibitors targeting Venezuelan equine encephalitis virus capsid protein identified using in silico structure-based-drug-design. Scientific Reports, 7(1), 1–6. doi: 10.1038/s41598-017-17672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, V. K., Srivastava, R., Sen Gupta, P. S., Naaz, F., Chaurasia, H., Mishra, R., Rana, M. K., & Singh, R. K. (2020). Anti-HIV potential of diarylpyrimidine derivatives as non-nucleoside reverse transcriptase inhibitors: Design, synthesis, docking, TOPKAT analysis and molecular dynamics simulations. Journal of Biomolecular Structure and Dynamics, 38, 1–8. 10.1080/07391102.2020.1748111 [DOI] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. Jan 30 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff, K. M., Rawlinson, S. M., Hearps, A. C., & Jans, D. A. (2011). An AlphaScreen®-based assay for high-throughput screening for specific inhibitors of nuclear import. Journal of Biomolecular Screening, 16(2), 192–200. Feb 10.1177/1087057110390360 [DOI] [PubMed] [Google Scholar]
- Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D., & Jans, D. A. (2012). Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. The Biochemical Journal, 443(3), 851–856. 10.1042/BJ20120150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li, C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu, Y., Maciejewski, A., Gale, N., Wilson, A., Chin, L., Cummings, R., Le, D., … Wilson, M. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research, 46(D1), D1074–D1082. 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica. B, 10(5), 766–788. Feb 27. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. N., Atkinson, S. C., Wang, C., Lee, A., Bogoyevitch, M. A., Borg, N. A., & Jans, D. A. (2020). The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Research, 177, 104760. DOI: 10.1016/j.antiviral.2020.104760 [DOI] [PubMed] [Google Scholar]
- Zhang, C., Zheng, W., Huang, X., Bell, E. W., Zhou, X., & Zhang, Y. (2020). Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. Journal of Proteome Research, 19(4), 1351–1360. Mar 22 10.1021/acs.jproteome.0c00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A Novel Coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.