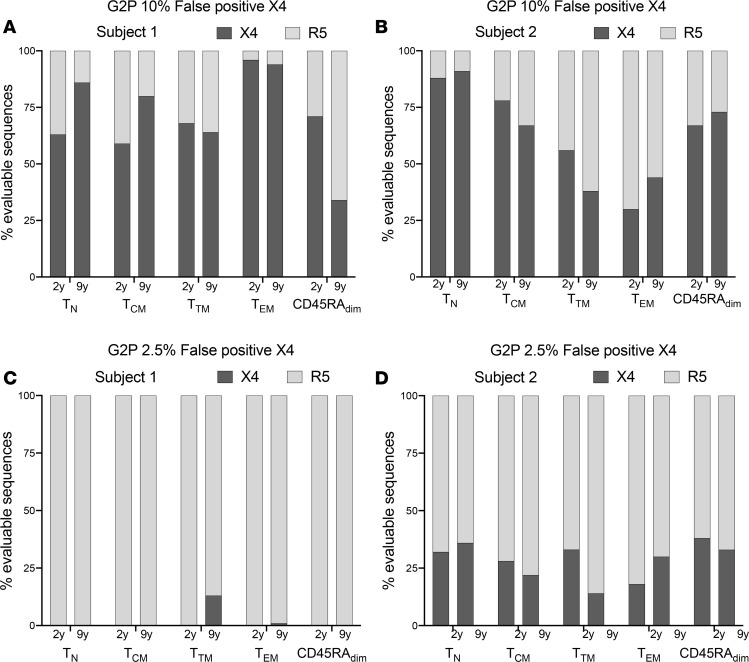
Figure 6. Analysis of coreceptor tropism using Geno2Pheno (G2P) suggests that TN cells might harbor CCR5-tropic HIV.
We used G2P to study coreceptor tropism of proviral sequences retrieved from different subsets. We included all the evaluable env sequences. (A and B) We used G2P with a 10% FPR, where the FPR indicates the likelihood to misclassify a provirus as CXCR4-tropic (which is based on phenotypic assays). When using G2P FPR 10%, both subjects showed a predominance of CXCR4-tropic sequences. (C and D) We used G2P with a 2.5% FPR. With G2P FPR 2.5%, most evaluable env ORFs were predicted to be CCR5-tropic for Subject 1 at both time points (C), while Subject 2 had, on average, 30% CXCR4-tropic sequences across subsets (D). Interestingly, with either 10% or 2.5% FPR, TN cells were predicted to contain CCR5-tropic sequences in both individuals.