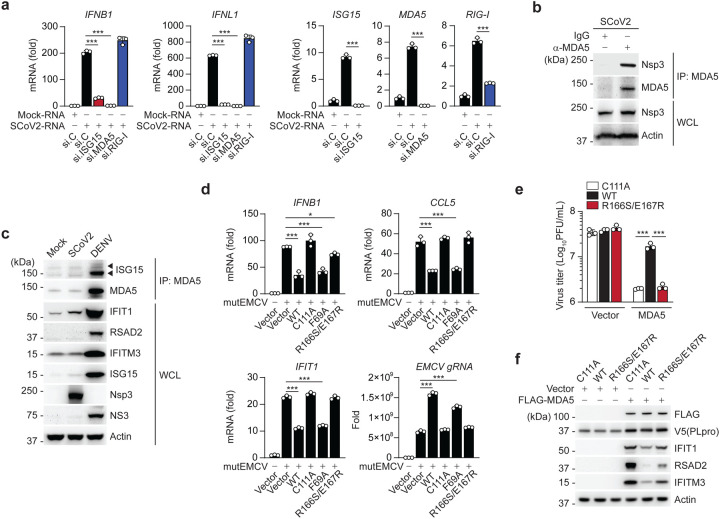
Figure 6. SCoV2 PLpro inhibits ISG15-mediated MDA5 signaling via its deISGylase activity.
(a) qRT-PCR analysis of IFNB1, IFNL1, ISG15, MDA5, and RIG-I transcripts in NHLFs that were transfected with the indicated siRNAs for 40 h and then transfected with mock-RNA or SCoV2-RNA (0.4 μg/mL) for 24 h. (b) Binding of SCoV2 Nsp3 to endogenous MDA5 in A549-hACE2 cells that were infected with SCoV2 (MOI 0.5) for 24 h, determined by IP with anti-MDA5 (or an IgG isotype control) followed by IB with anti-PLpro and anti-MDA5. WCLs were probed by IB with anti-PLpro (Nsp3) and anti-Actin. (c) Endogenous MDA5 ISGylation in A549-hACE2 cells that were mock-infected, or infected with SCoV2 or DENV (MOI 0.1 for each) for 48 h, determined by immunoprecipitation (IP) with anti-MDA5 followed by IB with anti-ISG15 and anti-MDA5. Protein abundance of IFIT1, RSAD2, IFITM3, ISG15 and actin in the WCLs were probed by IB. Efficient virus replication was verified by immunoblotting WCLs with anti-PLpro (Nsp3) or anti-NS3 (DENV). (d) qRT-PCR analysis of IFNB1, CCL5, IFIT1 transcript, and EMCV genomic RNA (gRNA) in HeLa cells that were transiently transfected for 24 h with vector, or V5-SCoV2 PLpro WT or mutants and then infected with mutEMCV (MOI 0.5) for 12 h. (e) EMCV titers in the supernatant of RIG-I KO HEK293 cells that were transiently transfected for 24 h with vector or FLAG-MDA5 along with V5-SCoV2 PLpro WT, C111A, or R166S/E167R and then infected with EMCV (MOI 0.001) for 16 h, determined by plaque assay. (f) Protein abundance of the indicated ISGs in the WCLs from the experiment in (e), determined by IB with the indicated antibodies. Data are representative of at least two independent experiments (mean ± s.d. of n = 3 biological replicates in a, d, e). *p < 0.05, ***p < 0.001 (unpaired Student’s t-test).