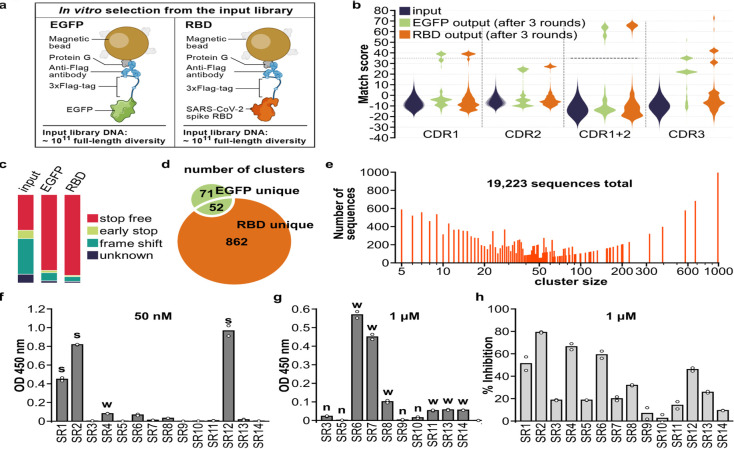
Fig. 2. Isolation and characterization of synthetic VHHs that binds SARS-CoV-2 spike RBD.
(a) Immobilization strategy for the target proteins: 3xFlag-tagged EGFP or RBD. (b) Pair-wise CDR match score (based on BLOSUM62 matrix) were calculated for 2000 randomly selected sequences from input library and output libraries after 3 rounds of selection. High match score populations appeared in the output libraries. Combining CDR1 and 2 match scores further separated high and low score population and a match score of 35 (black dashed line) was chosen as cut-off for downstream clustering analysis. (c) Percentage of indicated sequence categories in the input library and output libraries (EGFP, RBD). (d) Number of unique and shared clusters identified in EGFP and RBD output libraries. (e) Number of sequences for each size of RBD unique clusters. (f) ELISA assay revealed 3 strong binders (“s”) to RBD, 7 weak binders (“w”) and (g) 4 non-binders (“n”) among the 14 VHHs chosen for characterization. (h) SARS-CoV-2 S pseudotyped lentivirus neutralization assay showed 6 VHHs inhibiting infection >30% at 1μM on HEK293T expressing ACE2 and TMPRSS2. Data shown are two technical replicates, bars indicate the average of data, circles indicate values of each replicate.