SUMMARY
Many pathways of primary metabolism are substantially conserved within and across plant families. However, significant differences in organization and fluxes through a reaction network may occur, even between plants in closely related genera. Assessing and understanding these differences is key to appreciating metabolic diversity, and to attempts to engineer plant metabolism for higher crop yields and desired product profiles. To better understand lipid metabolism and seed oil synthesis in canola (Brassica napus), we have characterized four canola homologues of the Arabidopsis (Arabidopsis thaliana) ROD1 gene. AtROD1 encodes phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT), the enzyme that catalyzes a major flux of polyunsaturated fatty acids (PUFAs) in oil synthesis. Assays in yeast indicated that only two of the canola genes, BnROD1.A3 and BnROD1.C3, encode active isozymes of PDCT, and these genes are strongly expressed during the period of seed oil synthesis. Loss of expression of BnROD1.A3 and BnROD1.C3 in a double mutant, or by RNA interference, reduced the PUFA content of the oil to 26.6% compared with 32.5% in the wild type. These results indicate that ROD1 isozymes in canola are responsible for less than 20% of the PUFAs that accumulate in the seed oil compared with 40% in Arabidopsis. Our results demonstrate the care needed when translating results from a model species to crop plants.
Keywords: ROD1, fatty acids, seed lipid metabolism, metabolic diversity, seed oil, triacylglycerol
INTRODUCTION
Many plant species store triacylglycerols (TAGs) in their seeds as carbon and energy reserves to support germination and early seedling growth. Vegetable oil from crops such as canola (Brassica napus), soybean (Glycine max) and sunflower (Helianthus annuus) are a major source of calories in the human diet. Most vegetable oil TAGs contain differing proportions of five fatty acids including saturated (palmitic 16:0 and stearic 18:0), monounsaturated (oleic 18:1) and polyunsaturated (linoleic 18:2 and α–linolenic 18:3) acids (where the numerical abbreviations indicate carbon number:double bonds). The high proportion of polyunsaturated fatty acids (PUFAs) in commodity vegetable oils (35% and 60%, respectively, in traditional canola and soybean cultivars) pose a difficult problem for the food industry. This is because these levels of PUFAs greatly reduce the shelf life of products made from them, since 18:2 and particularly 18:3 oxidize on exposure to air and result in rancid off flavors (Weiss, 1983). Traditionally, these oils were stabilized by partial hydrogenation, designed to reduce the number of double bonds in the component fatty acids. However, the catalyst used to bring about the addition of hydrogen across double bonds also converts the natural cis double bonds to trans isomers (Weiss, 1983; Gurr, 1992). All-trans fatty acids behave biophysically (and thus physiologically) like saturated fatty acids. Furthermore, trans fatty acids are poorly metabolized by humans, so that trans fats are more damaging to human health than saturated fats (Steinhart et al., 2003; Micha and Mozaffarian, 2008, 2009; Wang et al., 2016). The recent US Food and Drug Administration rule banning trans fats from July 2018 has provided a compelling incentive to develop and grow crop cultivars with reduced levels of 18:2 and 18:3, with a concomitant increase in healthy monounsaturated 18:1 (Gillingham et al., 2011; Wilson, 2012; Wood et al., 2018; Bai et al., 2019). The ban on trans fats is based on studies indicating that they have contributed to thousands of unnecessary deaths a year in the USA (Micha and Mozaffarian, 2009; Wang et al., 2016). Successful reduction of polyunsaturates and increasing of monounsaturates in any oilseed will require detailed understanding of the pathways and enzymology of fatty acid metabolism and TAG synthesis in that plant species, because research indicates that different oilseeds use different combinations of pathways to synthesize their TAGs (Bates and Browse, 2012).
In developing oilseeds, fatty acids are synthesized in plastids and are exported into the cytosol mainly as 18:1 together with smaller amounts of 16:0 and 18:0 (Ohlrogge and Browse, 1995). Further modification of 18:1 occurs on the endoplasmic reticulum (ER). The dominant flux of 18:1 in many oilseeds, including canola, is to enter the membrane lipid phosphatidylcholine (PC) (Shanklin and Cahoon, 1998; Bates and Browse, 2012), where it can be desaturated by the ER-localized fatty acid desaturases FAD2 and FAD3 to produce 18:2 and 18:3 (Figure 1) (Miquel and Browse, 1992; Browse et al., 1993; Wallis et al., 2002). Thus, the ER oleate desaturase, FAD2, is the gateway enzyme for PUFA synthesis (Okuley et al., 1994). The resulting PUFAs may be transferred from PC to enter the acyl-CoA pool, or PUFA-rich PC may be converted to diacylglycerol by removal of the phosphocholine headgroup (Bates and Browse, 2012). Then, PUFA-rich TAGs may be produced from newly synthesized (‘de novo’) diacylglycerol (DAG) or PC-derived DAG (Bates and Browse, 2012), and PUFA-CoA by the acyl-CoA:diacylglycerol acyltransferases (DGATs) (Hobbs et al., 1999; Zou et al., 1999). Alternatively, PUFAs may be directly transferred from PC onto DAG to form TAG by an acyl-CoA-independent phospholipid:diacylglycerol acyltransferase (PDAT) (Dahlqvist et al., 2000; Zhang et al., 2009).
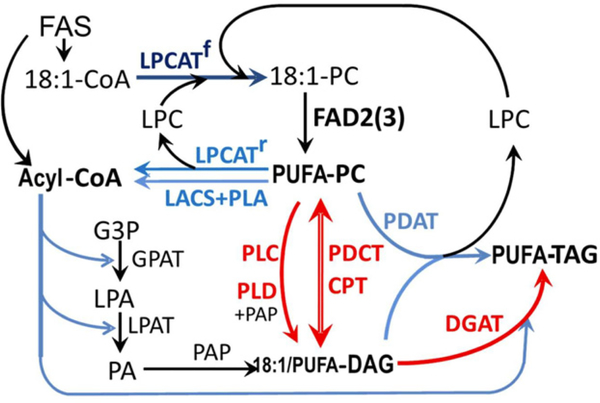
Figure 1. The metabolic network of reactions contributing to triacylglycerol (TAG) synthesis in oilseeds.
See text for details of pathway possibilities. The symmetrical phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) reaction that is the subject of this paper is shown by the double-headed red arrow. Abbreviations: CPT, cholinephosphotransferase; DAG, diacylglycerol; DGAT, diacylglycerol acyltransferase; FAD, fatty acid desaturase; FAS, fatty acid synthase; G3P, glycerol-3-phosphate; GPAT, G3P acyltransferase; LACS, long-chain acyl-CoA synthetase; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; LPAT, lysophosphatidic acid acyltransferase; LPCAT, LPC acyltransferase; PA, phosphatidic acid; PAP, PA phosphatase; PC, phosphatidylcholine; PDAT, phospholipid:diacylglycerol acyltransferase; PDCT, phosphatidylcholine:diacylglycerol cholinephosphotransferase; PLA, phospholipase A; PLC, phospholipase C; PLD, phospholipase D; PUFA, polyunsaturated fatty acid.
This relatively simple metabolism is complicated by the different enzymes that are capable of transferring fatty acids into and from PC (Figure 1). In developing seeds (and also in leaf tissue), newly synthesized 18:1 from the plastids enters PC by a process of acyl editing. Acyl editing is a deacylation–reacylation cycle of PC which exchanges the fatty acid on PC with fatty acid in the acyl-CoA pool using forward and reverse reactions of lysophosphatidylcholine acyltransferase (LPCAT) (Stymne and Stobart, 1984; Bates et al., 2007, 2009, 2012, 2013). Through acyl editing, 18:1 can be incorporated into PC for desaturation, and PUFAs can be released from PC to the acyl-CoA pool to be utilized along with 18:1-CoA for glycerolipid synthesis. Alternatively, 18:1 may enter PC through de novo glycerolipid synthesis (Figure 1) (Kennedy, 1961). This involves the sequential acylation of glycerol-3-phosphate (G3P) at the sn-1 and sn-2 positions to produce phosphatidic acid (PA) and subsequent removal of the phosphate group at the sn-3 position of PA by PA phosphatase (PAP) to produce de novo DAG. This DAG may be converted to PC by CDP-choline:diacylglycerol cholinephosphotransferase (CPT) (Slack et al., 1983; Goode and Dewey, 1999), as it is in most eukaryotes. It is also possible for PUFA-DAG to be derived from PC by the reverse activity of CPT (Slack et al., 1983, 1985). However, it is now clear that in the model plant Arabidopsis (Arabidopsis thaliana) the PC to DAG interconversion is catalyzed predominantly by a phosphatidyl choline:diacylglycerol cholinephosphotransferase (PDCT) through phosphocholine headgroup exchange between PC and DAG (Figure 1) (Lu et al., 2009). In Arabidopsis, PDCT is encoded by the AtROD1 gene (At3g15820) and genetic analysis indicates that the enzyme is responsible for 40% of the PUFA flux into TAG (Lu et al., 2009). Other enzymatic routes that may contribute to PUFA flux from the site of synthesis on PC to the DAG and acyl-CoA pools include phospholipase C, phospholipase D (+PAP) and phospholipase A (+long-chain acyl-CoA synthetase, LACS), and the action of these enzymes is also shown in Figure 1. Nevertheless, a recent study of lpcat1, lpcat2 and rod1 mutants indicates that in developing seeds of Arabidopsis these three enzymes account for at least 65% of the PUFA flux from PC into storage TAG, with the major flux being through PDCT (Bates et al., 2012).
In the context of these results for developing Arabidopsis seeds, we have investigated four BnROD1 genes from canola that are homologues of AtROD1. Assays of recombinant BnROD1 proteins in yeast (Saccharomyces cerevisiae) revealed two that encoded active PDCT isozymes, and we then identified mutations in the genes encoding these isozymes that eliminated enzyme activity. Our characterization of canola plants that contain these mutations suggests that PDCT in this oilseed crop is responsible for a considerably lower proportion of PUFA flux into TAG than is the case in Arabidopsis.
RESULTS
Transcript profiling of four B. napus ROD1 genes
Using the AtROD1 protein sequence (At3g15280) as the starting query, we carried out a series of BLAST (Altschul et al., 1990) database searches that identified four close homologues encoded in the allopolyploid B. napus genome (see Experimental procedures for details). Two genes in the A subgenome (derived from Brassica rapa) designated BnROD1.A3 and BnROD1.A5, where the nomenclature indicates the subgenome (A or C) and the chromosomal location of the gene (Chalhoub et al., 2014), encode isoforms with GenBank entries XP013736317 and XP013700801, respectively. Two genes in the C subgenome (derived from Brassica oleracea) designated BnROD1.C3 and BnROD1.C5 encode isoforms with GenBank entries XP013702685 and XP013695400, respectively. These four proteins are closely related to each other, exhibiting 86–98% sequence identity. Each of them shows approximately 80% identity to the AtROD1 protein (Table S1). Homologues of ROD1 are also found in other oilseeds; a cladogram showing the phylogenetic relationships among the canola proteins and those of other oilseed species is shown in Figure S1 in the online Supporting Information. An additional four B. napus genes that are more distantly related to AtROD1 (55–75% protein-sequence identity) were also identified, but these are not candidates for encoding PDCT activity in developing seeds (see Table S2).
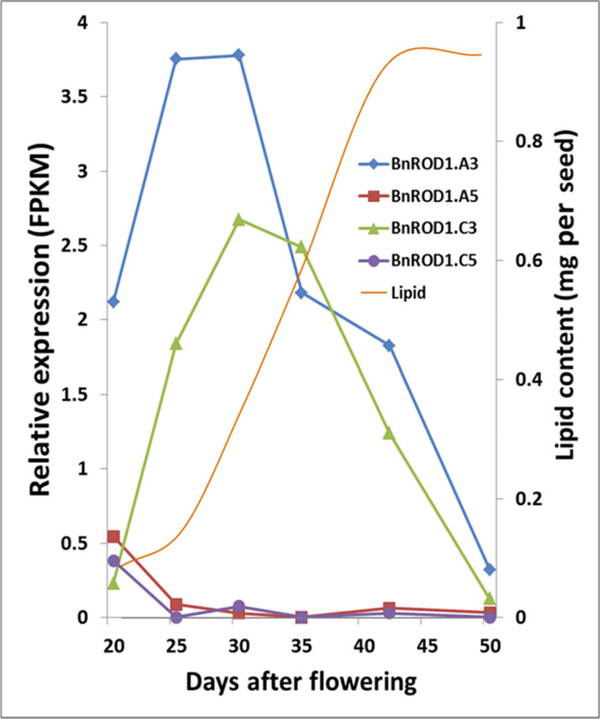
Transcript profiles of developing B. napus seeds were analyzed using established methods (Troncoso-Ponce et al., 2011) to determine expression of the four BnROD1 genes at six time points over the course of seed development. The data indicated that two of the genes, BnROD1.A3 and BnROD1.C3, exhibited a rapid increase in expression during early seed development, peaking at approximately 30 days after pollination, before gradually declining through the period of seed maturation up to 50 days after pollination (Figure 2). By contrast, both BnROD1.A5 and BnROD1.C5 were expressed at low to negligible levels throughout seed development. The strong expression of the BnROD1.A3 and BnROD1.C3 genes overlaps with the period of rapid TAG accumulation, which extends from 20 to 40 days after pollination (Perry and Harwood, 1993; Borisjuk et al., 2013). The extremely low expression of the BnROD1.A5 and BnROD1.C5 isoforms during the period of oil synthesis was confirmed in subsequent transcript-profiling experiments (Table S3).
Figure 2. Expression of BnROD1 genes during seed filling in Brassica napus.
Data are from RNA sequencing analyses of seed harvested at six stages of development. The course of oil accumulation is indicated by the orange line (right axis scale). FPKM, reads per kilobase per million reads in the database.
Expression of B. napus ROD1.A3 or BnROD1.C3 complements the Arabidopsis rod1 mutation
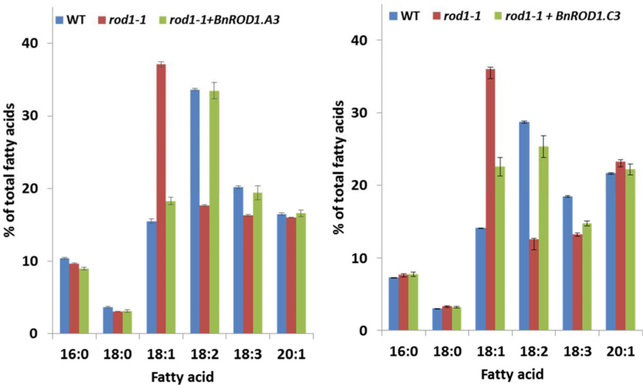
To first test whether the BnROD1.A3 gene encodes an active PDCT enzyme, we cloned a full-length cDNA and placed it under control of the seed-specific phaseolin promoter (Sengupta-Gopalan et al., 1985) in the pOEA2 transformation vector that allows identification of transgenic seed through the expression of the DsRed fluorescent protein (Lu et al., 2006). When we transformed mutant Arabidopsis rod1 plants, the T1 red seeds produced showed a reduction in 18:1 relative to the rod1 parent. We grew T1 plants to maturity, along with wild-type controls. Several of the T1 plants yielded T2 seed in approximate ratios of three red to one brown, indicating Mendelian segregation of a single DsRed/BnROD1.A3 transgene insert. For one of these, rod1;BnROD1.A3#4, we separated nine samples each of red and brown seed and analyzed FA compositions by gas chromatography. Compared with the wild-type control samples, the brown seeds had increased 18:1 and decreased 18:2 and 18:3 typical of rod1 seeds (Lu et al., 2009). By contrast, the red seeds had a distribution of 18-carbon fatty acids similar to the wild-type controls (Figure 3a), indicating that BnROD1.A3 cDNA encodes an active PDCT enzyme.
Figure 3. BnROD1.A3 and BnROD1.C3 complement an Arabidopsis rod1 mutant.
Data are seed fatty acid compositions from plants of wild type (WT), the rod1–1 mutant and rod1–1 expressing either a BnROD1.A3 (a) or BnROD1.C3 (b) transgene. Means ± SE (n = 9). Statistical principal component analysis indicates that the WT and each complemented line are highly similar and very different from the rod1–1 mutant.
Expression of the BnROD1.C3 coding sequence in the Arabidopsis rod1 mutant similarly resulted in substantial complementation of the rod1 seed fatty acid composition (Figure 3b), indicating that this gene also encodes an active PDCT. Statistical analysis of the data in Figure 3 by principal component analysis indicates that the wild type and each complemented line are highly similar and very different from the rod1–1 mutant.
In vitro assays of PDCT activity
To directly test the potential PDCT activity of the four BnROD1 isoforms, we placed the coding sequence of AtROD1 and each of the B. napus homologues under control of a yeast constitutive promoter in the vector p424GPD and expressed the proteins in strain HJ091 (cpt1::LEU2 ept1−), which lacks CPT activity (Morash et al., 1994). Microsomes were prepared from yeast expressing each construct and assayed for PDCT activity (Lu et al., 2009), based on conversion of [14C-glycerol]-diolein to PC.
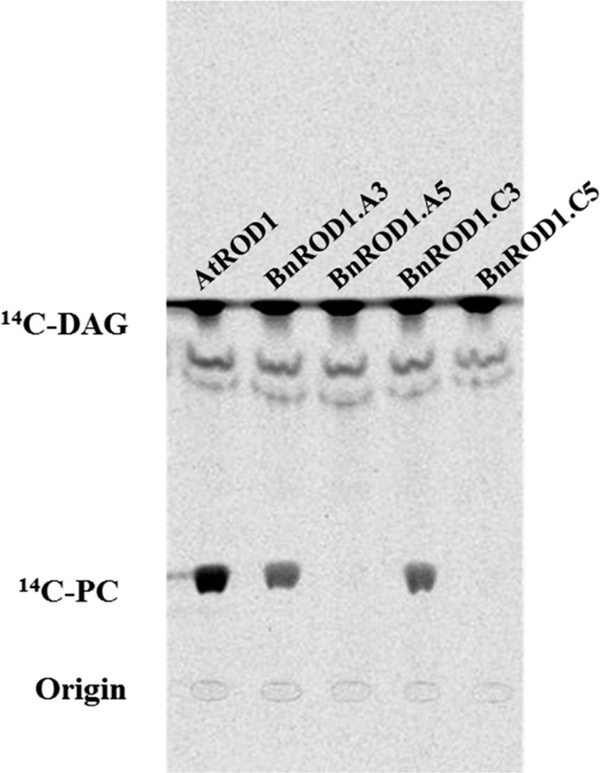
In these assays, microsomes from yeast expressing either BnROD1.A3 or BnROD1.C3 synthesized 14C-PC from the radioactive DAG substrate, like the AtROD1 control (Figure 4). However, no 14C-PC synthesis was detectable for microsomes from the yeast expressing BnROD1.A5 or BnROD1.C5 isoforms. These results indicate that there are only two isozymes of PDCT in B. napus, encoded by the BnROD1.A3 and BnROD1.C3 genes.
Figure 4.
Two BnROD1 genes encode active phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) in microsomes from yeast cells. Complementary DNAs of AtROD1 and four BnROD1 isogenes were expressed in yeast. Synthesis of 14C-phosphatidylcholine (14C-PC) from 14C-diacylglycerol (14C-DAG) demonstrates PDCT activity. Lipid extracts from yeast microsomes were separated by thin-layer chromatography and visualized in a phosphorimager.
Inhibition of BnROD1 expression by RNA-interference
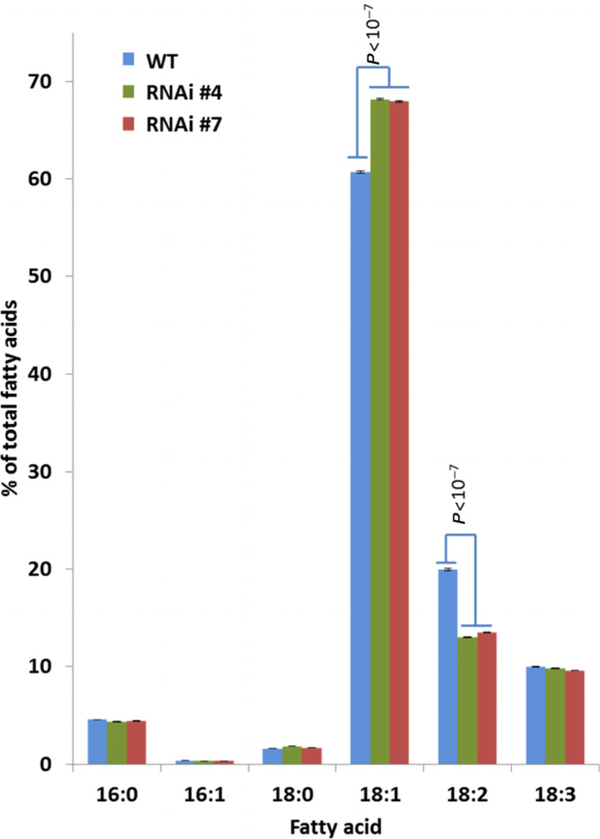
To assess the impact of the BnROD1 genes on lipid metabolism and FA composition in canola seeds, we designed a hairpin RNA construct to induce RNA interference (RNAi) against the BnROD1.A3 and BnROD1.C3 genes. This construct was based on PCR amplification of 278 bp of cDNA corresponding to the region from 29 bp to 300 bp of the BnROD1.A3 open reading frame. This sequence is 94% identical to the corresponding region of the BnROD1.C3 open reading frame with long sections of 100% identity, so the hairpin construct likely induces RNAi against both of these genes. By contrast, the sequence contains no region of homology greater than 20 bp with either the BnROD1.A5 or BnROD1.C5 genes. The 278-bp fragment from BnROD1.A3 was cloned into the pHELLSGATE12 vector (Wesley et al., 2001) to generate a hairpin construct, then the entire construct was subcloned into a transformation vector under control of the CaMV 35S promoter. Canola plants were transformed using a hypocotyl transformation protocol (De Block et al., 1989) and nine transgenic plants were recovered by selection for BASTA resistance encoded by the vector. The 18:1 content of seed samples from these nine T1 plants ranged from values similar to the wild type (58%) up to about 65% (Figure S2). Two T1 plants with high 18:1 were identified by pedigree analysis as having single RNAi transgene inserts. Homozygous progeny of these were grown to maturity along with untransformed parental controls. The FA compositions of mature T3 seed from these two lines, designated Bnrod1.a3/c3-RNAi#4 and Bnrod1.a3/c3-RNAi#7 are shown in Figure 5. Compared with the seed FA profile of the untransformed parental line, both RNAi lines showed a significant increase in 18:1 from 61% to 68% of the total fatty acids and reductions in the proportion of 18:2. These differences are smaller than those found for Arabidopsis rod1 seeds compared with the wild type (Figure 3) (Lu et al., 2009).
Figure 5. Expression of an RNA interference (RNAi) construct targeting BnROD1.A3 and BnROD1.C3 in seeds increases 18:1 content compared with the wild type (WT).
The fatty acid compositions of WT and two independent RNAi lines are shown. Means ± SE (n = 5). Statistical analyses (t-test; P-values shown) indicate highly significant differences in 18:1 and 18:2 between the RNAi lines and WT.
We also made a second hairpin construct based on the region of the BnROD1.A3 open reading frame from 289 bp to 849 bp. This 561-bp fragment shares substantial stretches of sequence identity with the other three BnROD1 transcript sequences and is likely to induce RNAi against all four genes. The hairpin construct was made and eight T1 transgenic canola plants were recovered as described above. The 18:1 content of seed samples from these plants was also increased, relative to wild-type controls (Figure S2), but we did not identify any lines with larger changes in 18:1 than had been produced by the hairpin construct designed to target only the BnROD1.A3 and BnROD1.C3 genes. This finding was not unexpected, since BnROD1.A5 and BnROD1.C5 have extremely low expression in developing seeds (Figure 2) and appear not to encode active PDCT enzyme (Figure 4).
Identification and characterization of mutants in the BnROD1.A3 and BnROD1.C3 genes
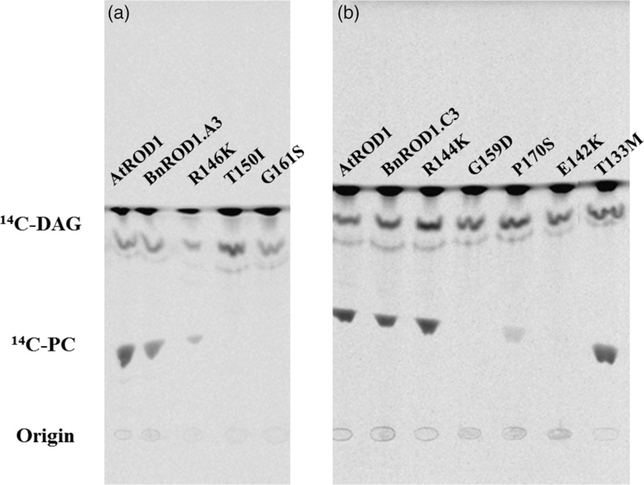
We next identified a series of mutations in the BnROD1.A3 and BnROD1.C3 genes from a sequence database of lines derived from a population of ethyl methanesulfonate (EMS) mutagenized canola plants. Initially, we identified three mutants in BnROD1.A3 that encode the amino acid changes R146K, T150I and G161S, and five mutants in BnROD1.C3 that encode the amino acid changes T133M, E142K, R144K, G159D and P170S. These mutations are indicated in the sequence alignment of ROD1 proteins shown in Figure S3.
To assess the effects of each mutation on the PDCT activity of the encoded protein, we used reverse transcriptase PCR and site-directed mutagenesis to obtain DNA corresponding to each of the eight mutant genes. These were cloned into the yeast expression vector p424GPD and, after confirmation of the sequence, expressed in yeast strain HJ091. Microsomes were prepared from cultures expressing the eight mutant proteins and wild-type controls and assayed for PDCT activity as described above. Both the R146K and T150I variants of the BnROD1.A3 protein showed low but detectable activity in the assay (about 14% and 3% of the wild-type protein, respectively) but the G161S protein had no detectable activity (Figure 6a). We designated this mutant line Bnrod1.a3–1. For the ROD1.C3 variants, R144K and T133M did not show any reduction in PDCT activity, while P170S retained approximately 15% of wild-type activity and E142K approximately 4%. Only the G159D variant had no detectable activity in the assay (Figure 6b).
Figure 6. Assays of phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) activity in yeast microsomes.
Assays of phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) activity in yeast microsomes for mutants of (a) BnROD1.A3 and (b) BnROD1.C3. Methods are as described in the caption to Figure 4. 14C-PC, 14C-phosphatidylcholine; 14C-DAG, 14C-diacylglycerol.
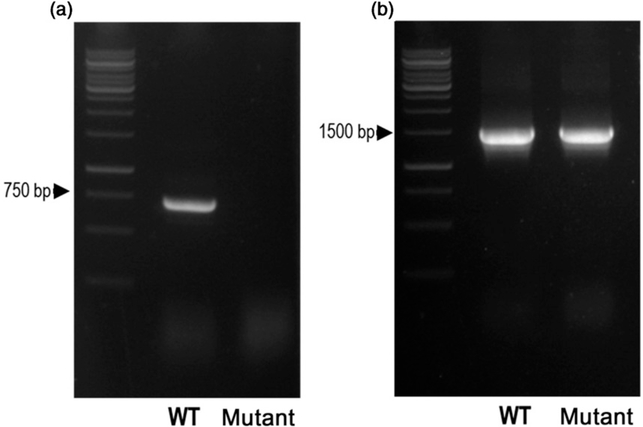
Unfortunately, the canola line carrying the PDCT G159D mutation had a poor growth phenotype that was not improved by two backcrosses to the wild type. It is possible that this phenotype is caused by a mutation tightly linked to the BnROD1.C3 gene. Further investigation of the mutant database identified a mutant line with a G to A transition at the donor site (CCA CAG|GT) of the second intron of BnROD1.C3, resulting in a downstream, in-frame TGA stop codon in the mutant sequence. To find out whether this mutation prevented expression of the gene, we prepared RNA from wild-type and mutant developing seeds and conducted RT-PCR, using primers spanning the two introns present in the coding sequence. Using RNA from the wild type yielded a product of about 700 bp corresponding to the size expected from the BnROD1.C3 transcript. However, RNA from mutant seeds did not yield any product, either of this size or of a size that would be expected if the second intron were retained (Figure 7a). Amplification of the BnDGAT1 transcript (Figure 7b) indicates that the RNA preparations from the two lines are of similar quality. We interpret these results as indicating that the mutation in the intron donor site leads to an aberrant product that is subject to degradation by RNA quality control (Elvira-Matelot et al., 2016). Thus, this line is a PDCT null, but growth of the plants is wild type. We designated this line Bnrod1.c3–1.
Figure 7. BnROD1.C3 is not expressed in a Bnrod1.c3–1 mutant line.
(a) Using RNA from developing seeds, primers detect BnROD1.C3 transcript in the wild type (WT) but not the Bnrod1.c3 mutant line. (b) Amplification of the BnDGAT1 transcript indicates RNA preparations are of similar quality. The left lane in each panel shows molecular weight markers. Expected RT-PCR product sizes are 728 bp for BnRod1.C3 and 1486 bp for BnDGAT1.
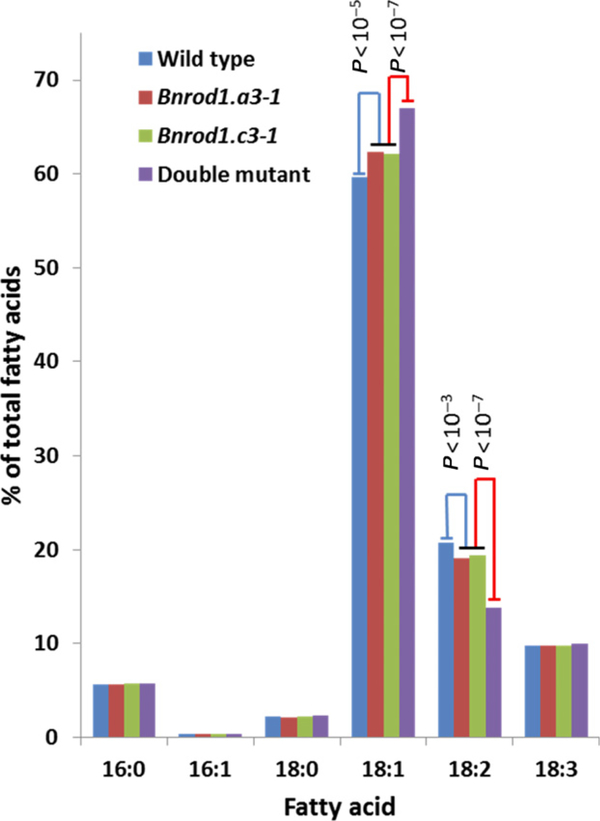
We crossed the two mutant lines, selfed the F1 progeny and from the F2 identified homozygous wild type, Bnrod1.a3–1, Bnrod1.c3–1 and double-mutant progeny by sequencing the two BnROD1 loci. When plants of these four lines were grown in a greenhouse, the single- and double-mutant plants were indistinguishable from the wild type in appearance. Although at this stage we did not analyze seed traits in these plants in detail, the mutants produced abundant seed that appeared normal. The FA composition of seed from each of the lines was determined by gas chromatography and is shown in Figure 8. Compared with the wild-type controls, each of the single mutants had a significant increase in the proportion of 18:1 in the seed oil, and a corresponding decrease in 18:2. In the double mutant these FA changes were larger, and comparable to the changes observed in the Bnrod1-RNAi lines (Figure 5). Taken together, these data indicate that canola deficient in PDCT activity show changes in seed oil composition that are similar to those observed in the Arabidopsis rod1 mutants (Lu et al., 2009), but smaller in magnitude.
Figure 8. Increased 18:1 content in Bnrod1 mutants.
Average seed fatty acid compositions for five samples each of wild type (WT), the Bnrod1.a3–1 and Bnrod1.c3–1 mutants, and a Bnrod1.a3–1/c3–1 double mutant grown in a greenhouse. All SE values (<0.5%) were too small to show. Statistical analyses (t-test; P-values shown) indicate highly significant differences in 18:1 and 18:2 between each single mutant and WT, and between the double mutant and each single mutant.
Field evaluation of the Bnrod1 mutants
To complete our evaluation of the growth and oil characteristics of canola lines deficient in PDCT activity, we grew our single- and double-mutant lines together with wild-type segregants at three field sites with three replicates at each site. Each plot of plants was evaluated during growth using 10-point assessment scales for seedling establishment, plant vigor at bolting and maturity. Time from emergence to the beginning and end of flowering (10% and 90% of plants with flowers, respectively) was also recorded. Seed samples from each plot were analyzed for oil, protein and glucosinolate content, and the FA composition of the oil was determined by gas chromatography. The data obtained for these agronomic and seed traits were assessed by anova to calculate the least significant difference (LSD) for each parameter.
Results for growth and seed components of the four lines are shown in Table 1. Comparison of the data for the single- and double-mutant plants with the wild type showed only small increases or decreases in the range of 2–5% change, and none of these is statistically significant. However, data for the seed FA compositions in these field-grown plants (Table 2) show statistically significant changes for all three of the mutant lines relative to wild-type controls, similar to those found for greenhouse-grown plants (Figure 8), with an increase in the proportion of 18:1 and decrease in 18:2. In the Bnrod1.a3–1 Bnrod1.c3–1 double-mutant seed, 18:1 was 63.5% of total fatty acids compared with 57.4% in the wild type, while 18:2 decreased from 22.7% in wild-type seeds to 16.9% in the double mutant (Table 2). In addition this data set reveals a small, but statistically significant, decrease in 16:0 for the three mutant lines.
Table 1.
Growth and physiology of Brassica napus lines with mutations in Bnrod 1 genes during field trials. ANOVA analysis was used to calculate the least significant difference (LSD) values
| Genotype | Establishment | vigor | Days to flowering | End of flowering | Maturity | Protein (%) | Glucosinolates (μmol g−1) | Oil (%) |
|---|---|---|---|---|---|---|---|---|
| Wild type | 4.9 | 4.9 | 42 | 65.5 | 4.8 | 25.3 | 8 | 43.7 |
| Bnrod1.a3–1 | 4.8 | 4.5 | 41.5 | 64.8 | 5.1 | 24.1 | 7.8 | 45.2 |
| Bnrod1.c3–1 | 4.9 | 4.8 | 41.4 | 63.9 | 4.8 | 25.3 | 9.6 | 44.5 |
| Double mutant | 4.8 | 5 | 41.4 | 63.8 | 4.6 | 24.5 | 8.4 | 44.9 |
| LSD | 0.4 | 0.6 | 1.7 | 2.5 | 0.7 | 2.3 | 2.5 | 2.9 |
Table 2.
Seed fatty acid compositions of Brassica napus lines with mutations in Bnrod 1 genes, harvested from field trials. ANOVA analysis was used to calculate least significant difference (LSD) values
| Genotype | Percentage of total fatty acids |
||||
|---|---|---|---|---|---|
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | |
| Wild type | 4.8* | 1.6* | 57.4* | 22.7* | 9.8 |
| Bnrod1a3–1 | 4.5* | 1.7* | 59.4* | 21.4* | 9.5 |
| Bnrod1.c3–1 | 4.5* | 1.7* | 59.6* | 21.0* | 9.6 |
| Double mutant | 4.6* | 1.8* | 63.5* | 16.9* | 9.6 |
| LSD | 0.1 | 0.1 | 1.4 | 1.1 | 0.6 |
Asterisks indicate values that are significantly different (P < 0.05) from the wild type, based on ANOVA analysis.
DISCUSSION
Across the range of angiosperm species that synthesize oil reserves in their seeds there are substantial variations in the selection of enzymes and pathways used for TAG synthesis. The network of possible paths from FA synthesis in the plastids to TAG localized in cytoplasmic oil bodies is summarized in Figure 1. A review of 19 different oilseeds (Bates and Browse, 2012) compared species that predominantly desaturate (or otherwise modify) fatty acids on PC by reactions in the ER and then rely on PC-derived DAG for TAG synthesis with species that use de novo DAG to synthesize TAG without diversion into the ER PC pool. Canola, other Brassica species and Arabidopsis fall into the middle of the continuum between these two extremes (Bates and Browse, 2012).
For species such as canola and Arabidopsis that rely quite heavily on PC-derived DAG there are further choices about the reactions that can mobilize DAG, and fatty acids, into PC and from PC into TAG synthesis. For PC to DAG conversion, PDCT, CPT, phospholipase D and phospholipase C (+PAP) are all possibilities (Figure 1). In addition, LPCAT, PDAT and PLA (+LACS) are routes by which fatty acids at sn2 (and possibly sn1) of PC can be mobilized for TAG synthesis. The Brassica oilseeds, including canola, and camelina (Camelina sativa) are close relatives of Arabidopsis, and many aspects of TAG synthesis, and seed biochemistry more broadly, have been found to be similar (Kelly et al., 2013; Aznar-Moreno et al., 2015; Jiang et al., 2017). However, the complexity of the metabolic network shown in Figure 1 underscores the value and importance of investigating and evaluating the contributions of particular enzymes and reactions within the network to the flux of fatty acids and glycerol towards TAG.
In this study we have employed biochemical and genetic approaches to identify two genes, BnROD1.A3 and BnROD1.C3, that encode the isozymes providing PDCT activity for TAG synthesis in developing canola seeds. Two additional homologous genes, BnROD1.A5 and BnROD1.C5, appear not to contribute to PDCT in seeds. All four of the proteins encoded by these genes were identified as having approximately 80% identity and 88% similarity to the Arabidopsis PDCT enzyme (Table S1). Transcript profiling data indicate that both BnROD1.A3 and BnROD1.C3 have a peak of expression in developing seeds (Figure 2) that correlates with the period of TAG accumulation, and recombinant proteins showed PDCT activity when expressed in yeast (Figure 4). We also expressed BnROD1.A3 and BnROD1.C3 in the Arabidopsis PDCT mutant rod1, and complemented the high 18:1 phenotype of this mutant (Figure 3). By contrast, recombinant BnROD1.A5 and BnROD1.C5 proteins did not have PDCT activity in our yeast assays (Figure 4) and neither of these genes has any substantial expression during the period of oil synthesis in seeds (Figure 2).
We used our yeast assay to measure the relative activities of variants of BnROD1.A3 and BnROD1.C3 that are encoded by mutant genes identified in a screen of canola lines from a population derived from EMS treatment. Two mutant lines, designated Bnrod1.a3–1 (G161S) and Bnrod1.c3–1 (an intron-splice mutant), that are predicted null alleles were selected for characterization. After back-crossing each mutant to the wild type at least three times, we crossed the two mutants, selfed the F1 progeny plants and from the population of F2 plants selected homozygous wild-type, Bnrod1.a3–1, Bnrod1.c3–1 and double-mutant segregants. Analyses of seed FA compositions in each of the single mutants showed increases in the proportion of 18:1 and decreases in 18:2 relative to wild-type segregants, and changes of greater magnitude were observed in the double mutants. The changes in FA composition were similar for plants grown in a greenhouse (Figure 8) and those grown in our field trials (Table 2), although some quantitative differences in FA composition were observed. These are likely due to differences in temperature, light and other environmental conditions between the greenhouse and the field sites. Changes in 18:1 and 18:2 in the double-mutant seeds were similar to those seen in seeds of plants in which BnROD1.A3 and BnROD1.C3 were targeted by RNAi (Figure 5). As we observed in our characterization of rod1 mutant Arabidopsis (Lu et al., 2009), the elimination of PDCT activity had no measurable effect on growth and development of canola plants (Table 1).
A difference between canola and Arabidopsis is the quantitative effects of PDCT deficiency on seed FA composition. In seeds of our double-mutant canola, the products of 18:1 desaturation, 18:2 + 18:3, are reduced from 32.5% in the wild type to 26.6%. Arabidopsis wild-type seeds contained 49.1% 18:2 + 18:3 and this was lowered to 29.4% in seeds of the rod1 mutant, and from these data we calculated that 40% [100 × (49.1 – 29.4)/49.1] of the 18:1 that is converted to 18:2 and 18:3 enters PC via the PDCT enzyme (Lu et al., 2009). The same calculation using our data from wild-type and PDCT-deficient canola seed indicates that only 18.2% of 18:1 enters PC via PDCT. This finding indicates that despite the close phylogenetic relationship between Arabidopsis and canola, the crop species uses a complement of fluxes through the network of TAG synthesis pathways shown in Figure 1 that differs from that used by the model species Arabidopsis.
In this study we have used biochemical and genetic approaches to thoroughly investigate the enzymatic and metabolic functions of canola PDCT and to assess the role of this enzyme in the flux of fatty acids into and from PC during fatty acid desaturation and TAG synthesis in developing canola seeds. One unexpected finding from our experiments is that, in canola, PDCT has a considerably smaller role in converting DAG to PC (with the simultaneous conversion of PC to DAG) than is the case in the model oilseed Arabidopsis. Nevertheless, eliminating PDCT activity does have a modest effect in reducing PUFA levels in canola oil – from 32.5% to 26.6% in our field trials – without negatively affecting the growth and development of the plants. These findings indicate that the Bnrod1 mutants may be useful in efforts to reduce PUFAs in plant oils, and the consequent production of trans fats in processed shortenings and food products made from them.
EXPERIMENTAL PROCEDURES
Identification and expression analysis of B. napus ROD1 genes
A TBLASTN homology search using the AtROD1 protein sequence (At3g15280) as the query resulted in the identification of the Unigene sequence Bna.6194 from a B. napus expressed sequence tag database as the homologue of AtROD1. Subsequently, Bna.6194 was used as the query in a BLAST homology search of BASF proprietary databases of B. rapa coding sequences and of B. oleracea coding sequences. The contigs in these databases were obtained by assembly of short sequence reads using the software package SOAPdenovo. The BLAST analyses resulted in the identification of four candidate genes for B. rapa and four for B. oleracea. BLAST homology searches of a proprietary BASF database containing B. napus mRNA sequences using the B. rapa and B. oleracea gene sequences, and a TBLASTN homology search of the gene database for B. napus cultivar Darmor using the AtROD1 protein sequence, followed by gene structure predictions using the FGENESH software and RNA sequencing (RNAseq) read abundance, resulted in the identification of the cDNA sequences of four possible B. napus ROD1 genes, two from the A subgenome and two from the C subgenome. These sequences encode peptides with GenBank entries XP013736317, XP013700801, XP013702685 and XP013695400, here designated BnROD1.A3, BnROD1.A5, BnROD1.C3 and BnROD1.C5, respectively (Chalhoub et al., 2014). The four more distantly related sequences (GenBank entries XP013676101, XP013682366, XP013714642 and XP013717241) contain amino acid changes and expression patterns that indicate that they are not candidates for encoding PDCT activity in developing seeds (see notes in Table S2).
The relative gene expression levels of the four B. napus ROD1 genes were determined through analysis of Illumina RNAseq-derived transcriptome data obtained for six different seed developmental stages, using established methods (Troncoso-Ponce et al., 2011). Gene expression levels were calculated (Figure 2) taking into account a normalization step for the sequencing depth per database (target reads per million reads in the database) and for the target gene length (reads per kilobase per million reads in the database, FPKM) (Mortazavi et al., 2008), or (Table S2) as counts per transcript (transcripts per million, tpm) determined as described by Li and Dewey (2011).
Complementation of the Arabidopsis rod1 mutant
Transformation of Arabidopsis rod1–1 plants with a cDNA of BnROD1.A3 under control of the seed-specific phaseolin promoter was carried out as described by Lu et al. (2009). Seeds of A. thaliana ecotype Col-0 (wild type), the rod1–1 mutant and the rod1;BnROD1.A3#4 line (rod1–1 expressing the BnROD1 cDNA) were sown directly on soil. The sown seeds were incubated at 5°C for 48 h, then cultivated at 22°C with 16 h light at 100–150 μE m−2. Seeds from mature plants were collected for analysis. Transformation of rod1–1 with a cDNA of BnROD1.C3 was performed in the same way and the FA compositions of T1 seeds was analyzed.
Fatty acid and lipid analysis
The overall FA composition of Arabidopsis seeds was determined by heating samples at 80°C in 1 ml of 2.5% (v/v) H2SO4 in methanol and a known amount of 17:0 internal standard for 1.5 h in screw-capped tubes. After the addition of 1.5 ml of 0.9% NaCl solution and 1 ml of hexane, fatty acids were extracted into the hexane phase by shaking and the tubes were centrifuged at low speed. Samples (1 μl) of the hexane phase were separated by gas chromatography on a 15 m × 0.53 mm Carbowax column and quantified using a flame ionization detector. The gas chromatograph was programmed for an initial temperature of 150°C for 3 min followed by an increase of 15°C min−1 to 210°C; this final temperature was maintained for a further 12 min.
For analysis of canola seed samples, about 0.8 g of seed (ground or whole) was weighed and added to 2.0 ml of sodium methoxide solution, 1.0 ml of petroleum ether and a metal crushing rod in a scintillation vial and crushed in an Eberbach shaker to extract the oil. The fatty acids of the O-acyl lipids were converted to their methyl ester derivatives by sodium methoxide and extracted into the hexane phase. These methyl esters were analyzed by capillary gas–liquid chromatography on a Zebron ZB-Wax column (15 m × 0.32 mm ID × 0.50 μm film thickness) in a Varian Model 430 chromatograph, with a flame ionization detector.
Yeast expression and enzyme assays
A cDNA for each BnROD1 gene was obtained by reverse transcription PCR using RNA isolated from wild-type plants using primers that introduced BamH1 and EcoR1 restriction sites. Following BamHI and EcoRI double digestion, each product was ligated into the p424GPD vector (ATCC, http://www.atcc.org/), in which the cDNA is expressed under control of the constitutive glyceraldehyde-3-P dehydrogenase promoter, and then transformed into Escherichia coli competent cells (TOP10, Invitrogen). Plasmids with correct inserts confirmed by sequencing were transformed into yeast HJ091 yeast cells (cpt1::LEU2 ept1–), and transformants were selected by growth on synthetic minimal medium (SD base) with dropout leucine and tryptophan (DO–Leu/–Trp) (Clontech, https://www.clontech.com). Microsomes prepared from the yeast cultures were used to assay for PDCT activity as described by Lu et al. (2009). To obtain cDNAs corresponding to the sequences in Bnrod1 mutant plants, a wild-type BnROD1.A3 or BnROD1.C3 sequence was used as a template for alteration using a Phusion site-directed mutagenesis kit (Invitrogen, https://www.invitrogen.com). The resulting mutated cDNA was cloned and its sequence confirmed before expression in yeast and assay of PDCT activity as described above.
Suppression of BnROD1 genes by RNA interference
To specifically knock down the expression of BnROD1.A3 and BnROD1.C3, a hairpin construct was generated. A 271-bp fragment from bp 29–300 of BnROD1 was amplified by PCR using primers BnROD1 F1 (bp 29–53) CACCGTCGCAGATCTAACGGATATCACAC (forward) and BnROD1 R1 (bp 276–300) AATATCGAACGGCTCAGACTTCGCC (reverse). A 561-bp fragment from bp 289–849 of BnROD1 was amplified by PCR using primers BnROD1 F2 CGTTCGATATTGGGTTTGTGGCCACGCGC (forward) and BnROD1 R2 GGCTCAGACTTCGCCGGAACCATCTGGAGC (reverse). The DNA fragments were amplified by PCR on BnROD1.A3 DNA as a template. The PCR reaction (50 μl) contained 0.3 μmol of each primer, 2 ng μl−1 of template DNA, 0.2 mM of dNTP mix, 0.02 unit μl−1 of KOD DNA polymerase (Toyobo, https://www.toyobo-global.com/), 5 μl of 10× PCR buffer and 1.5 mM MgSO4. Programmed cycles were as follows: 2 min initial denaturing step at 95°C; 40 cycles of 20 sec denaturation at 95°C, 15 sec annealing at 55°C, 20 sec extension at 70°C. The PCR products were purified with QIAquick Gel Extraction Kit (Qiagen) and ligated into the pENTR.™/D-TOPO.R™ cloning vector (Invitrogen) to generate entry clones according to the instruction manual. To generate hairpin constructs, 100 ng of BnROD1 entry clone and 150 ng of pHELLSGATE12 destination vector were mixed and an LR recombination reaction was conducted using Gateway™ LR Clonase enzyme, following the instructions (Invitrogen). After transformation into TOP10 competent cells, clones were screened by restriction analysis to identify plasmids with the expected insert in the correct orientation, and were then validated by sequencing.
Transformation vectors were obtained by extracting the hairpin region from the above constructs and placing this cassette into a transformation vector under control of the CaMV 35S promoter. This vector was used for transformation of B. napus plants using the hypocotyl transformation protocol as described by De Block et al. (1989). Single-copy regenerated B. napus transformation events were back-crossed to the wild type. After selfing of the resulting progeny, seeds from both homozygous transformation events and wild-type segregants were harvested and analyzed for FA composition as described above.
Generation and isolation of mutant B. napus rod1 alleles
Mutations in the BnROD1 genes were generated and identified as follows. 30 000 seeds from an elite spring oilseed rape breeding line were pre-imbibed for 2 h on wet filter paper in deionized water. Half of the seeds were exposed to 0.8% EMS and half to 1% EMS (Sigma M0880, https://www.sigmaaldrich.com/) and incubated for 4 h. The mutagenized seeds (M1 seeds) were rinsed three times and dried in a fume hood overnight. M1 plants were grown in soil and selfed to generate M2 seeds. M2 seeds were harvested for each individual M1 plant. A total of 2 × 4800 M2 plants, derived from different M1 plants, were grown and DNA samples were prepared from leaf samples of each individual M2 plant. The DNA samples were screened for the presence of point mutations in the ROD1 genes by direct sequencing and the sequences were analyzed for the presence of point mutations using NovoSNP software (Weckx et al., 2005; http://www.molgen.ua.ac.be/bioinfo/novosnp/). The mutant lines selected for detailed investigation were backcrossed to the wild type at least three times before being used in the reported experiments.
To assess the expression of the Bnrod1.c3–1 mutation, RNA was extracted from developing B. napus seeds using the Qiagen RNeasy Plant Mini Kit (cat no./ID 74904) following the manufacturer’s instructions. Qiagen DNase was used for on-column DNA digestion following the manufacturer’s instructions. The transcript was amplified using the Thermofisher SuperScript IV One-Step PCR System (catalog no. 12594025, https://www.thermofisher.com/), according to the manufacturer’s instructions. Primers used for BnROD1.C3 were: GGCAACGGAGTAGGAGGGAAGAGCAAGGCGTCG (forward) and GATTAATTGACTAGCGAGTCTTTAGAAATCAAACTAAAACCATTGGC (reverse). For BnDGAT1: ATGGAGATTTTGGATTCTGGAGGCGTCACTATGC (forward) and CTATGACATCTTTCCTTTGCGGTTCATCAAGTCGTGAT (reverse).
Oil composition in seeds from Bnrod1 mutants grown in the greenhouse
Plants of the Bnrod1.a3–1 and Bnrod1.c3–1 lines were crossed. Following selfing of the resulting progeny, seeds from plants homozygous for Bnrod1.a3 or Bnrod1.c3 mutations, the double mutant and wild-type segregants were identified by sequencing of the two loci. The FA composition of the seed oil of these lines was determined by gas chromatography, as described above.
Measurements of plant growth and seed composition of mutant lines in the field
Fatty acid composition and plant performance parameters were determined for plants grown in the field. Brassica napus lines homozygous for Bnrod1.a3 or Bnrod1.c3 mutations, the double mutant and wild-type segregants were grown at three different geographical locations, west and south of Ghent, Belgium at (1) Astene, (2) Kruishoutem and (3) Volkegem. The sites had different soil types (1, sandy; 2, sand + low loam; 3, sand + high loam) but similar seasonal weather patterns. At the time of planting (21–22 May 2012), night-time minimum temperatures were close to the long-term averages (10–11°C). At each site, plants of each line were grown in four replicate 2 m × 1.5 m plots with six rows of plants in each plot. During most of the 4-month growing season, average diurnal maximum and minimum temperatures were in the ranges 17–22°C and 10–14°C, respectively. Rainfall over the 4-month growing season at the sites was approximately 240 mm (1), 200 mm (2) and 180 mm (3). Harvest dates were 12 September (1) and 7 September (2 and 3).
The FA composition of the seed oil was determined as described above. The following plant performance parameters were determined: establishment at the two to three leaf stage on a scale of 1–9, where 1 is very thin, 5 is average and 9 is very thick; vigor at the five to six leaf stage on a scale of 1–9, where 1 is poor, 5 is average and 9 is vigorous; flowering start, the stage (in days after seeding) at which 10% were in flower; flowering end, the stage (in days after seeding) at which 10% remained in flower; maturity on a scale of 1–9, where 1 is late, 5 is average and 9 is early; glucosinolate content (as μmol N g−1); oil content in the seed (as a percentage of the whole seed); protein content in the seed (as a percentage of the whole seed). Seed FA compositions were obtained by gas chromatography. For the statistical analysis an anova test was performed to identify significant differences between the mutant lines and the wild-type segregants.
Statistical analyses
Student’s t-test was used to evaluate the FA compositions shown in Figures 5 and 8. For the complementation experiments in Figure 3 we undertook principal component analysis using the prcomp function in R (Venables and Ripley, 2013). This function uses singular value decomposition of the FA compositions of the lines. The calculations were performed using RStudio software. For the multisite field experiments, anova was used to calculate the LSD for each of the parameters measured, with a criterion of P < 0.05 indicating statistically significant differences.
Supplementary Material
Table S1. Pair-wise percentage identity and similarity between Brassica napus ROD1 proteins and Arabidopsis ROD1 (AtROD1).
Table S2. Pair-wise percentage identity and similarity between additional proteins and Arabidopsis ROD1 (AtROD1).
Table S3. Expression of four BnROD1 genes at six time-points during seed filling in Brassica napus.
Figure S1. Cladogram showing the implied phylogeny of eight ROD1 proteins.
Figure S2. Seed 18:1 content in 17 RNA interference lines.
Figure S3. Protein sequence alignment of phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) enzymes from Brassica napus (BnROD1.A3 and BnROD1.C3) and Arabidopsis (AtROD1).
ACKNOWLEDGEMENTS
This work was supported by BASF Innovation Center Gent, Agriculture Food Research Initiative (AFRI) competitive award number 2018-67013-27459 from the USDA National Institute of Food and Agriculture and by the Agricultural Research Center at Washington State University. We are grateful to Jessica Coppens and Mieke Stevens who were involved in the cloning of the B. napus ROD1 genes, to Chris Opsomer who generated the hairpin constructs, to Liesbeth Vercruysse and Lieven Paermentier for growing and molecular characterization of the plants, to Phil Bates for suggesting the yeast assays of PDCT and to our colleagues at BASF Innovation Center Gent and WSU for helpful discussion.
Footnotes
CONFLICT OF INTERESTS
The authors declare no competing financial interests.
DATA AVAILABILITY STATEMENT
Original data from experiments reported in this paper have been archived on the Research Exchange at Washington State University and are available at https://research.libraries.wsu.edu/xmlui/handle/2376/16922.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ (1990) Basic local alignment search tool. J. Mol. Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Aznar-Moreno J, Denolf P, Van Audenhove K, De Bodt S, Engelen S, Fahy D, Wallis JG and Browse J (2015) Type 1 Diacylglycerol Acyltransferases of Brassica napus preferentially incorporate oleic acid into triacylglycerol. J. Exp. Bot 66, 6497–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Engelen S, Denolf P, Wallis JG, Lynch K, Bengtsson JD, Van Thournout M, Haesendonckx B and Browse J (2019) Identification characterization and field testing of Brassica napus mutants producing high-oleic oils. Plant J 98, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD and Browse J (2012) The Significance of different diacylglycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci 3, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Ohlrogge JB and Pollard M (2007) Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J. Biol. Chem 282, 31206–31216. [DOI] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB and Pollard M (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 150, 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Fatihi A, Snapp AR, Carlsson AS, Browse J and Lu C (2012) Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol 160, 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Stymne S and Ohlrogge J (2013) Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol 16, 358–364. [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Neuberger T, Schwender J et al. (2013) Seed architecture shapes embryo metabolism in oilseed rape. Plant Cell, 25,1625–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, McConn M, James D Jr and Miquel M (1993) Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem 268,16345–16351. [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S et al. (2014) Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H and Stymne S (2000) Phospholipid : diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA, 97, 6487–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block M, De Brouwer D and Tenning P (1989) Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the Expression of the bar and neo genes in the transgenic plants. Plant Physiol 91,694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira-Matelot E, Bardou F, Ariel F, Jauvion V, Bouteiller N, Le Masson I, Cao J and Crespi MD (2016) The nuclear Ribonucleoprotein SmD1 interplays with splicing, RNA quality control, and posttranscriptional gene silencing in Arabidopsis. Plant Cell, 28, 426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham LG, Harris-Janz S and Jones PJ (2011) Dietary monounsaturated fatty acids are protective against metabolic syndrome and cardiovascular disease risk factors. Lipids, 46, 209–228. [DOI] [PubMed] [Google Scholar]
- Goode JH and Dewey RE (1999) Characterization of aminoalcoholphosphotransferases from Arabidopsis thaliana and soybean. Plant Physiol. Biochem 37, 445–457. [Google Scholar]
- Gurr MI (1992) Role of Fats in Food and Nutrition, 2nd edn London: Elsevier Applied Science. [Google Scholar]
- Hobbs DH, Lu C and Hills MJ (1999) Cloning of a cDNA encoding diacylglycerol acyltransferase from Arabidopsis thaliana and its functional expression. FEBS Lett 452, 145–149. [DOI] [PubMed] [Google Scholar]
- Jiang WZ, Henry IM, Lynagh PG, Comai L, Cahoon EB and Weeks DP (2017) Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J 15, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Shaw E, Powers SJ, Kurup S and Eastmond PJ (2013) Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol. J. 11, 355–361. [DOI] [PubMed] [Google Scholar]
- Kennedy EP (1961) Biosynthesis of complex lipids. Fed. Proc. Am. Soc. Exp. Biol 20, 934–940. [PubMed] [Google Scholar]
- Li B and Dewey C (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fulda M, Wallis JG and Browse J (2006) A High-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J 45, 847–856. [DOI] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M and Browse J (2009) An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc. Natl. Acad. Sci. USA, 106,18837–18842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micha R and Mozaffarian D (2008) Trans fatty acids: effects on cardiometabolic health and implications for policy. Prostaglandins Leukot. Esset. Fatty Acids, 79,147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micha R and Mozaffarian D (2009) Trans fatty acids: effects on metabolic syndrome, heart disease and diabetes. Nat. Rev. Endocrinol 5, 335–344. [DOI] [PubMed] [Google Scholar]
- Miquel M and Browse J (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J. Biol. Chem 267, 1502–1509. [PubMed] [Google Scholar]
- Morash SC, McMaster CR, Hjelmstad RH and Bell RM (1994) Studies employing Saccharomyces cerevisiae cpt1 and ept1 null mutants implicate the CPT1 gene in coordinate regulation of phospholipid biosynthesis. J. Biol. Chem 269, 28769–28776. [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L and Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J and Browse J (1995) Lipid Biosynthesis. Plant Cell, 7, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E and Browse J (1994) Arabidopsis Fad2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell, 6, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry HJ and Harwood JL (1993) Changes in lipid content of developing seeds of Brassica napus. Phytochemistry, 32,1411–1415. [Google Scholar]
- Sengupta-Gopalan C, Reichert NA, Barker RF, Hall TC and Kemp JD (1985) Developmentally regulated expression of the bean betaphaseolin gene in tobacco seed. Proc. Natl. Acad. Sci. USA, 82, 3320–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J and Cahoon EB (1998) Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol 49, 611–641. [DOI] [PubMed] [Google Scholar]
- Slack CR, Campbell LC, Browse JA and Roughan PG (1983) Some evidence for the reversibility of cholinephosphotransferase-catalyzed reaction in developing linseed cotyledons in vivo. Biochim. Biophys. Acta, 754, 10–20. [Google Scholar]
- Slack CR, Roughan PG, Browse JA and Gardiner SE (1985) Some properties of Cholinephosphotransferase from developing safflower cotyledons. Biochim. Biophys. Acta, 833, 438–448. [Google Scholar]
- Steinhart H, Rickert R and Winkler K (2003) Trans fatty acids (TFA): analysis, occurrence, intake and clinical relevance. Eur. J. Med. Res 8, 358–362. [PubMed] [Google Scholar]
- Stymne S and Stobart AK (1984) Evidence for the reversibility of the acyl-CoA:lysophosphatidylcholine acyltransferase in microsomal preparations from developing safflower (Carthamus tinctorius L) cotyledons and rat-liver. Biochem. J 223, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso-Ponce MA, Kilaru A, Cao X, Durrett TP, Fan J, Jensen JK, Thrower NA, Pauly M, Wilkerson C and Ohlrogge JB (2011) Comparative deep transcriptional profiling of four developing oilseeds. Plant J 68,1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN and Ripley BD (2013) Modern Applied Statistics with S-PLUS. Berlin, Heidelberg: Springer Science & Business Media. [Google Scholar]
- Wallis JG, Watts JL and Browse J (2002) Polyunsaturated fatty acid synthesis: what will they think of next? Trends Biochem. Sci 27, 467–473. [DOI] [PubMed] [Google Scholar]
- Wang DD, Li Y, Chiuve SE, Stampfer MJ, Manson JE, Rimm EB, Willett WC and Hu FB (2016) Association of specific dietary fats with total and cause-specific mortality. JAMA Intern. Med, 176, 1134–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckx S, Del-Favero J, Rademakers R, Claes L, Cruts M, De Jonghe P, Van Broeckhoven C and De Rijk P (2005) NovoSNP, a novel computational tool for sequence variation discovery. Genome Res 15, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss TJ (1983) Food Oils and Their Uses, 2nd edn Westport, CT: AVI Publishing. [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Wilson RF (2012) The role of genomics and biotechnology in achieving global food security for high-oleic vegetable oil. J. Oleo Sci. 61, 357–367. [DOI] [PubMed] [Google Scholar]
- Wood CC, Okada S, Taylor MC et al. (2018) Seed-specific RNAi in safflower generates a superhigh oleic oil with extended oxidative stability. Plant Biotech. J 16,1788–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC and Ohlrogge JB (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell, 21, 3885–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Wei Y, Jako C, Kumar A, Selvaraj G and Taylor DC (1999) The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J 19, 645–653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Pair-wise percentage identity and similarity between Brassica napus ROD1 proteins and Arabidopsis ROD1 (AtROD1).
Table S2. Pair-wise percentage identity and similarity between additional proteins and Arabidopsis ROD1 (AtROD1).
Table S3. Expression of four BnROD1 genes at six time-points during seed filling in Brassica napus.
Figure S1. Cladogram showing the implied phylogeny of eight ROD1 proteins.
Figure S2. Seed 18:1 content in 17 RNA interference lines.
Figure S3. Protein sequence alignment of phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) enzymes from Brassica napus (BnROD1.A3 and BnROD1.C3) and Arabidopsis (AtROD1).