Fig 6.
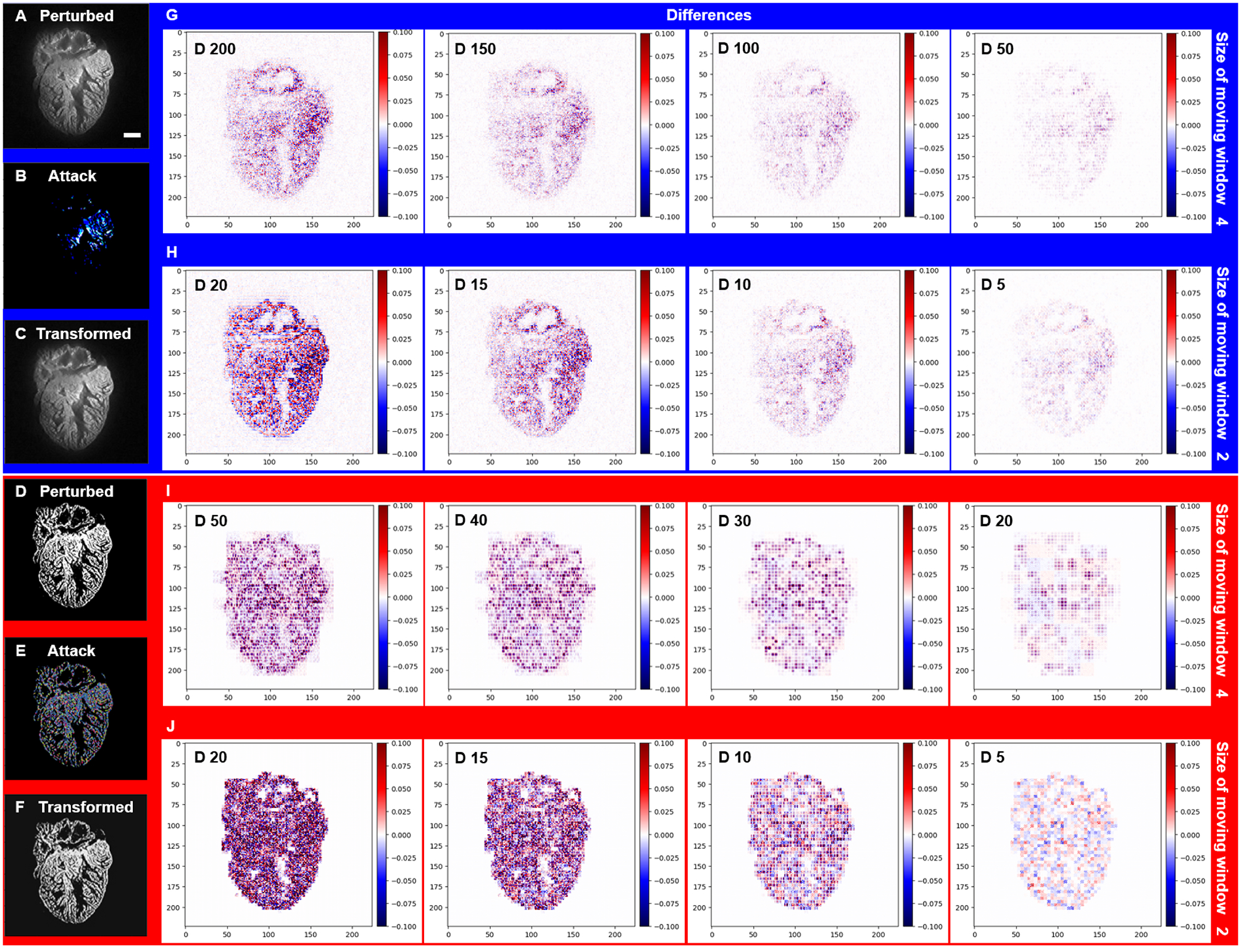
Forward and inverse Saak transforms to filter the adversarial perturbations from LSFM images. (A and D) Adversarial perturbed images were generated by Deepfool. (B and E) Perturbations were added to both grayscale and binary images. (C and F) Transformed images were reconstructed following truncation of the high-frequency coefficients. (G) By applying the 4 × 4 moving window, 200 (73% of total), 150 (55%), 100 (37%) and 50 (18%) coefficients (spectral dimensions) were filtered out from the grayscale image, respectively, and (I) 50 (18% of total), 40 (15%), 30 (11%) and 20 (7%) coefficients were filtered out from the binary image, respectively. (H and J) By applying the 2 × 2 moving window, 20 (95% of total), 15 (71%), 10 (48%) and 5 (24%) coefficients were filtered out from both the grayscale and binary images, yielding the representative differences from the binary image. The pseudo-color represents the intensity of specified adversarial perturbation. D stands for number of Saak coefficients. Scale bar: 200 μm.
