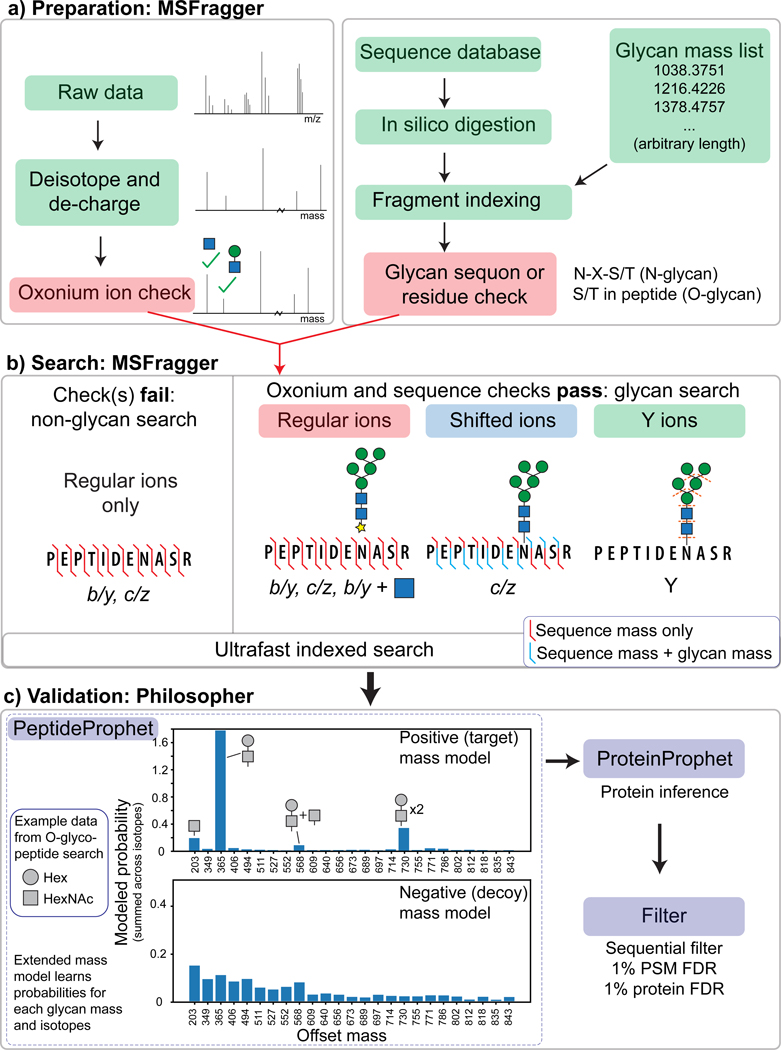
Figure 1.
Workflow of MSFragger-Glyco. a) Data and database preparation. Raw MS/MS spectra are processed (left) and searched against a protein database (right). Peptides containing a possible glycosite have additional glyco-specific fragments added to the index. b) Spectra are searched against indexed peptides. If the spectrum contains oxonium ion(s) and the peptide being considered contains a possible glycosite, a glycan search is performed (right); if either check fails, a regular search is performed. Shifted ions (blue) contain the intact mass of the glycan on the peptide while regular ions (red) contain only the masses of the amino acid residues. c) FDR filtering is performed using Philosopher. Plot of probabilities learned by PeptideProphet for a subset of mass offsets searched (mass 0 to 850 Da, masses with target probability >1% only) from O-glycopeptide data shows high probability for common compositions (e.g. HexNAc-Hex at 365) in the positive model, whereas the negative model shows similar probability for across mass shifts. Probabilities displayed are summed across isotope errors (0/1/2), resulting in probabilities than can exceed a value of 1.