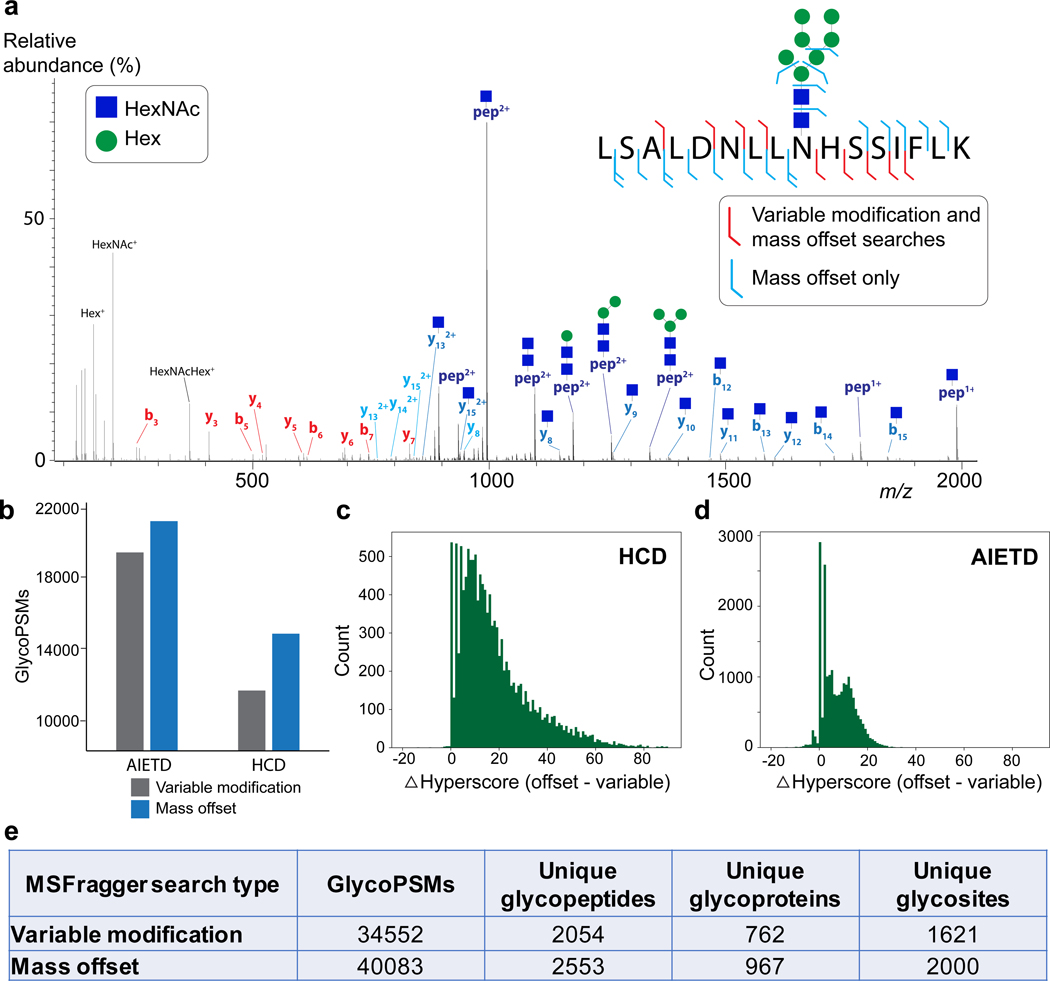
Figure 2.
Comparison of mass offset and variable modification-type searches for N-linked glycopeptides. a) Example HCD tandem mass spectrum of peptide LSALDNLLNHSSIFLK with glycan HexNAc2Hex7. Fragment ions that match the identification assuming the intact glycan is present at N-9 (variable modification-type) are colored red. Note that none of the fragments annotated in red contain the glycosite. Fragments in blue correspond to the mass offset search, including Y ions, b/y ions without the glycan or with a single HexNAc (blue square) remaining. Oxonium ions are shown in black (not all are labeled). b) Number of glycoPSMs obtained for MSFragger-Glyco mass offset search (orange) or variable modification search (blue) from AIETD and HCD spectra. c) Score difference (mass offset hyperscore – variable modification hyperscore) for spectra that were annotated in both search types for HCD spectra and (d) for AI-ETD spectra, showing a larger improvement in scores for HCD data. e) Table of results for MSFragger-Glyco mass offset and variable modification searches of 16 glycans.