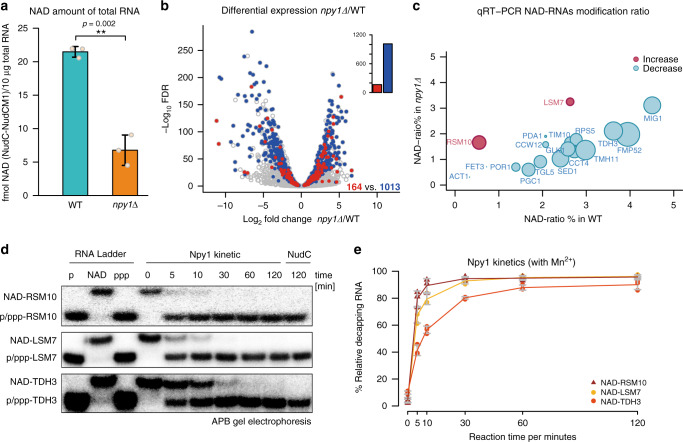
Fig. 4. RNAs are affected differently by Npy1 deletion.
a Quantification of NAD-RNA from total RNA by LC-MS. Integrated NAD content, which was determined via the NR signal intensity (fmol) from washed and NudC-treated total RNA (10 µg), while subtracting the NR signal intensity obtained from RNA treated with a NudC mutant. Dots represent individual biological triplicate measurements. Error bars represent mean ± SD. Performed in biologically independent replicates, n = 3. p-values are denoted by asterisks: **p < 0.01 (Student’s t-test, one-sided). b Volcano plot of transcripts comparing WT with npy1Δ samples by RNA expression level and NAD-modification ratio. The log2 fold change of transcript abundance from transcriptome sequencing (npy1Δ vs. WT) is plotted versus the log10 false discovery rate (FDR). Different transcripts (7620) were analyzed and represented as dots. Red dots represent RNAs for which the NAD ratio increased upon NPY1 gene deletion according to NAD captureSeq (NAD ratio: npy1Δ > WT > 0, transcriptome normalized base mean >100, p < 0.05, FDR < 0.1), and blue dots represent RNAs for which the NAD ratio decreased (NAD ratio: WT > npy1Δ > 0, transcriptome normalized base mean >100, p < 0.05, FDR < 0.1). c Bubble plot of the relative NAD-modification ratio of 18 RNA species, as determined by qRT-PCR. Blue bubbles represent RNAs for which the NAD-modification ratio decreased upon NPY1 gene deletion, whereas red bubbles show those with increased NAD-modification ratio. The bubble size indicates the extent of the relative change. d Npy1 in vitro kinetics of decapping different NAD-RNAs. NAD-RSM10 (5′-end fragment, 107 nt), NAD-LSM7 (5′-end fragment, 30 nt), and NAD-TDH3 (5′-end fragment, 99 nt) were assayed. All conditions as in Fig. 3a with the exception of a twofold higher enzyme concentration. Three independent experiments were performed, n = 3. e Quantification of Npy1-mediated 5′-NAD decapping over time, as shown in Fig. 4d. Error bars represent the mean ± SD. Three independent experiments were performed, n = 3. Source data are provided as a Source Data file.