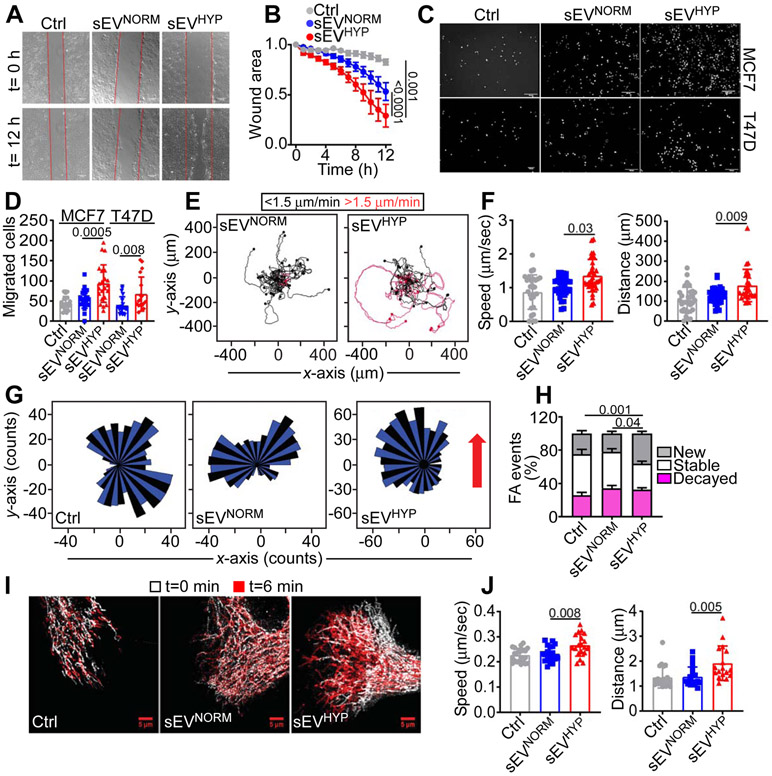
Figure 1. Breast cancer-derived hypoxic sEV (sEVHYP) stimulate motility of normal mammary epithelial cells.
(A) Normal mammary epithelial MCF10A cells treated with small extracellular vesicles (sEV) produced by MCF7 cells under normoxic (eEVNORM) or hypoxic (sEVHYP) conditions and analyzed for directional cell migration in a wound closure assay. Representative images at time (t) 0 h and 12 h. Scale bar, 150 μm.
(B) The conditions are as in (A) and directional motility of sEV-treated MCF10A recipient cells was quantified. Mean±SEM (N=5). Numbers correspond to p values by 2-way Anova with Tukey's multiple comparisons test.
(C and D) MCF10A cells treated with sEVNORM or sEVHYP produced by MCF7 (top) or T47D (bottom) cells were analyzed for migration on PET inserts (C, representative images of DAPI-stained nuclei of migrated cells) and quantified (D). Scale bar, 200 μm. Mean±SEM (N=3). Numbers correspond to p values by 1-way Anova with Tukey's posttest.
(E and F) MCF10A cultures treated with MCF7 cell-derived sEVNORM or sEVHYP were analyzed for cell motility in 2D contour plots (E) and the speed of cell movements (F, left) or total distance traveled (F, right) by individual sEV-treated MCF10A cells was quantified. Each tracing corresponds to an individual cell. The cutoff velocities for slow (black, <1.5 μm/min)- or fast (red, >1.5 μm/min)-moving cells are indicated. Mean±SD (N=3). Numbers correspond to p values by 1-way Anova with Tukey's posttest.
(G) MCF10A cells treated with MCF7 cell-derived sEVNORM or sEVHYP were analyzed for chemotactic cell motility quantified by rose plots. Red arrow, direction of the chemotactic gradient. (N=3).
(H) MCF10A cells treated with MCF7 cell-derived sEVNORM or sEVHYP were analyzed for focal adhesion (FA) turnover and the percentage of new, stable or decayed FA was quantified. Mean±SEM (N=3). Numbers correspond to p values by 2-way Anova with Tukey's posttest.
(I and J) MCF10A cells as in (H) were analyzed for mitochondrial motility by time-lapse videomicroscopy (I, representative images) at time (t) 0 and 6 min, and the speed of mitochondrial movements (J, left) and total distance traveled by individual mitochondria (J, right) was quantified. Mean±SD (N=3). Scale bar, 5 μm. Numbers correspond to p values by 1-way Anova with Tukey’s posttest.