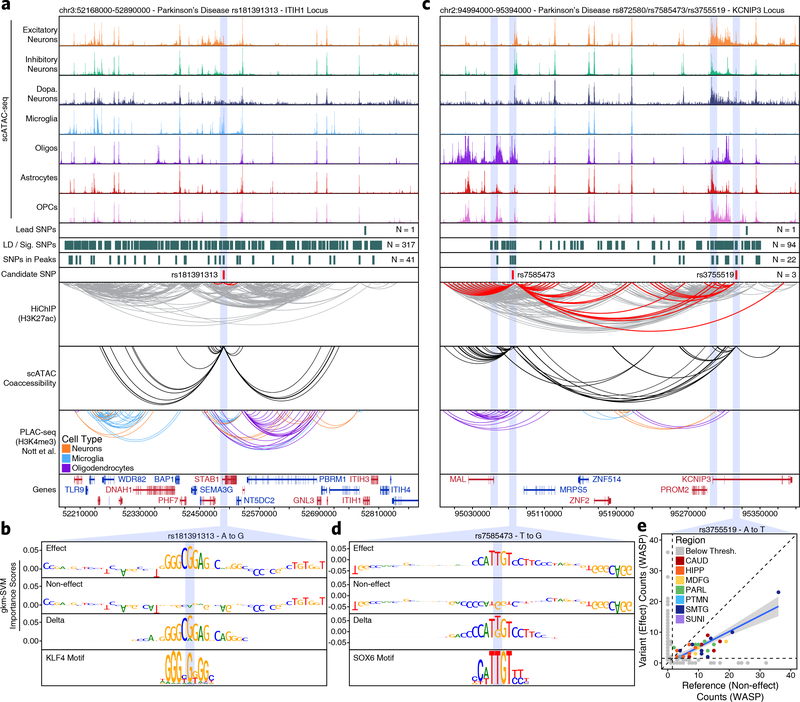
Fig. 4 – Vertical integration of multi-omic data and machine learning nominates gene targets in AD and PD.
a,c, Normalized scATAC-seq-derived pseudo-bulk tracks, H3K27ac HiChIP loop calls, co-accessibility correlations, and publically available H3K4me3 PLAC-seq loop calls (Nott et al. 2019) in (a) the ITIH1 gene locus (chr3:52168000–52890000) or (c) the KCNIP3 locus (chr2:94994000–95394000). scATAC-seq tracks represent the aggregate signal of all cells from the given cell type and have been normalized to the total number of reads in TSS regions. For HiChIP, each line represents a FitHiChIP loop call connecting the points on each end. Red lines contain one anchor overlapping the SNP of interest. For co-accessibility, only interactions involving the accessible chromatin region of interest are shown. For PLAC-seq, MAPS loop calls from microglia (blue), neurons (orange), and oligodendrocytes (purple) are shown. b,d, GkmExplain importance scores for each base in the 50-bp region surrounding (b) rs181391313 or (d) rs7585473 for the effect and non-effect alleles from the gkm-SVM model corresponding to (b) microglia (Cluster 24) or (d) oligodendrocytes (Cluster 21). The predicted motif affected by the SNP is shown at the bottom and the SNP of interest is highlighted in blue. e, Dot plot showing allelic imbalance at rs3755519. The bulk ATAC-seq counts for the reference/non-effect (A) allele and variant/effect (T) allele are plotted. Each dot represents an individual bulk ATAC-seq sample (N = 140) colored by brain region. Samples where fewer than 3 reads were present to support both the reference and variant allele (i.e. presumed homozygotes or samples with insufficient sequencing depth) are shown in grey. The blue line represents a linear regression of the non-grey points and the grey box represents the 95% confidence interval of that regression.