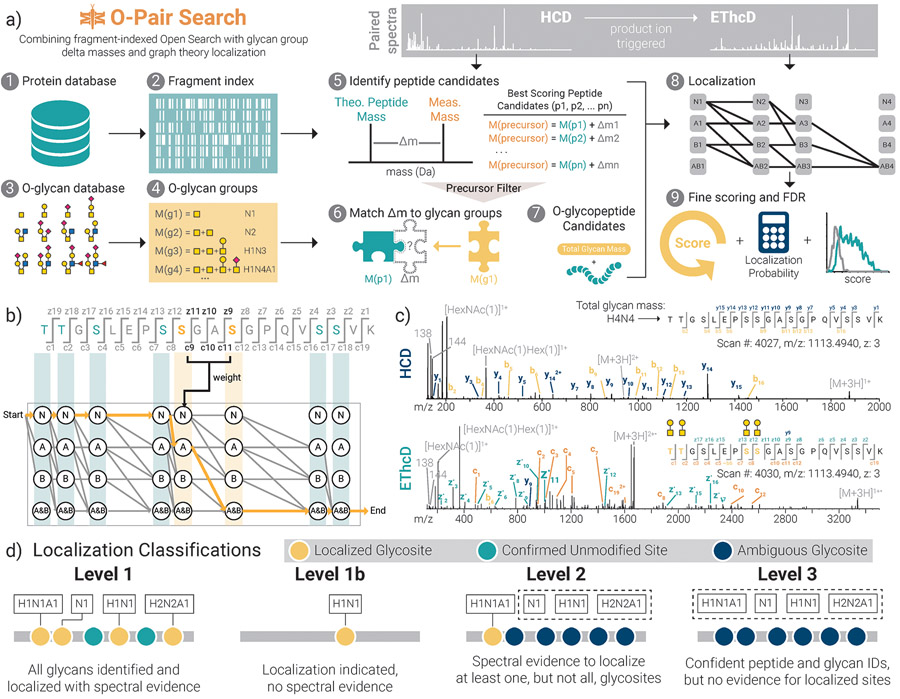
Figure 1. O-Pair Search through MetaMorpheus for fast and confident identification of O-glycopeptides.
a) The workflow describes processing steps in the O-Pair Search strategy, which generates a fragment ion index [1, 2] and O-glycan groups [3, 4] from user defined protein and O-glycan databases, respectively. Using an ultrafast, fragment-index-enabled open modification search [5] paired with a match of delta masses to aggregate glycan mass combinations [6] enables identification of O-glycopeptide candidates from HCD spectra [7]. Paired EThcD spectra are then used for graph theory-based localization calculations to rapidly assign modification sites for all glycans comprising the O-glycan group [8]. Finally, more detailed re-scoring of spectra, localization probability calculations, and false discovery rate corrections are performed before returning identifications to the user [9]. b) A demonstration of graph theory-based localization using a hypothetical example of an O-glycopeptide TTGSLEPSSGASGPQVSSVK from human mucin-type O-glycoprotein CD43 (leukosialin), which has 8 potential O-glycosites. Here we consider how graph theory determines O-glycosites using c/zdot fragments present in EThcD spectra when two glycans (termed A and B for the sake of demonstration) are presented as modifications. c) An example of paired HCD and EThcD spectra for quadruply-O-glycosylated TTGSLEPSSGASGPQVSSVK, showing a Level 1 identification where all calculated glycan mass shifts can be confidently localized to discrete residues. Note, no fragments in the HCD spectrum retain any glycan masses. Rather, the thorough peptide backbone fragmentation without glycan retention shows how the sequence was confidently retrieved with a defined mass shift matching a combination of O-glycans. The subsequent EThcD spectrum then enables localization of all 4 O-glycosites (gold) even with the presence of 4 other unmodified potential sites. d) O-Pair Search defines levels of localization for each GlycoPSM. Note, “H”, “N”, and “A” represent hexose, HexNAc, and Neu5Ac, respectively.