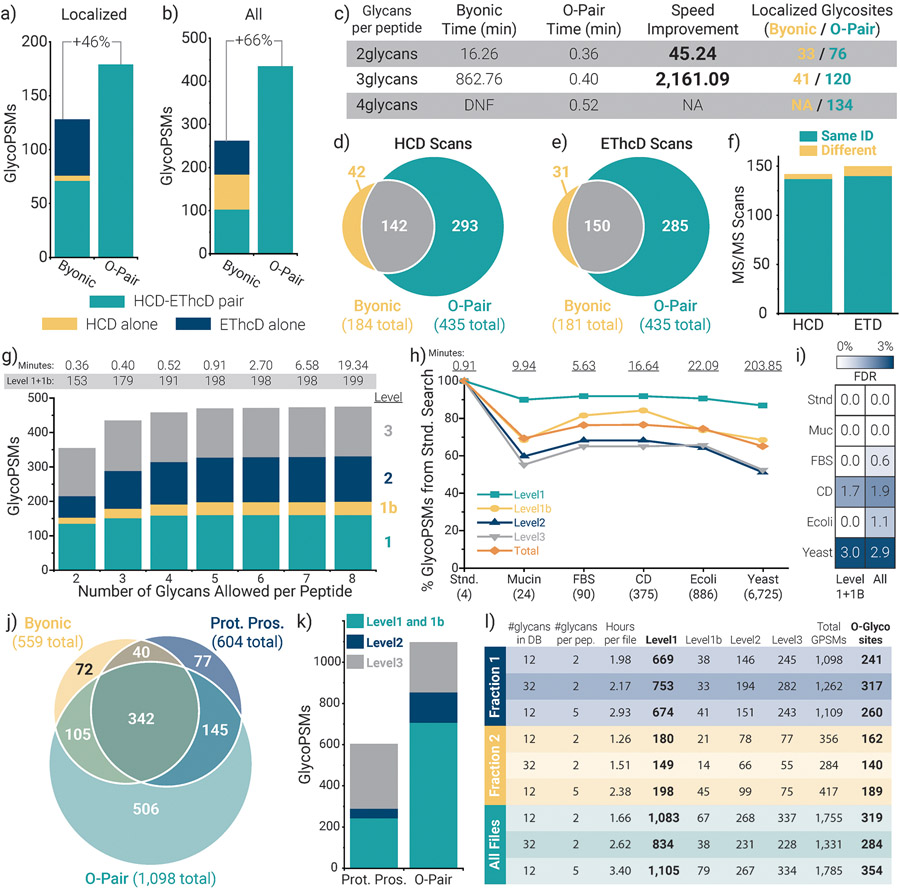
Figure 2. Performance of O-Pair Search for O-glycopeptide characterization.
Comparing the number of a) localized and b) total glycopeptide spectral matches (GlycoPSMs) returned from Byonic and from O-Pair Search for HCD-pd-EThcD data collected from StcE digestions of four recombinant mucin standards. Note, only Level 1 and 1b identifications are considered for the localized O-Pair Search data, and 3 glycans per peptide were allowed for both searches. c) The table compares the search times required for Byonic and O-Pair Search when considering 2, 3, and 4 glycans per peptide (DNF = did not finish). The number of localized glycosites identified by the searches is also provided, which correspond to the GlycoPSMs in panel a and Supplementary Fig. 3. Overlap in GlycoPSMs between O-Pair Search and Byonic is compared for both d) HCD and e) EThcD scans, and f) the majority (~95%) of the shared identified scans mapped to the same glycopeptide. g) O-Pair Search enabled consideration of more glycans per peptide while keeping search times reasonable. h) O-Pair Search also allowed the use of several different protein database backgrounds much larger in size without untenable search time increases. Here, “Total” indicates all identifications, i.e., the sum of all Localization Level identifications. i) Use of entrapment databases with proteins not present in the sample did not inflate false discovery rates above approximately 1-3%. j) O-Pair Search was used to process files from a published urinary O-glycopeptide study that previously reported Protein Prospector (Prot. Pros.) and Byonic results. k) Protein Prospector reports localized glycosites, which we converted into our Localization Level system and compared with O-Pair results. l) Results from several O-Pair searches of Fraction 1 (three files), Fraction 2 (two files), and all ten files available from the urinary O-glycopeptide study. Supplementary Note 4 details the files used for each panel.