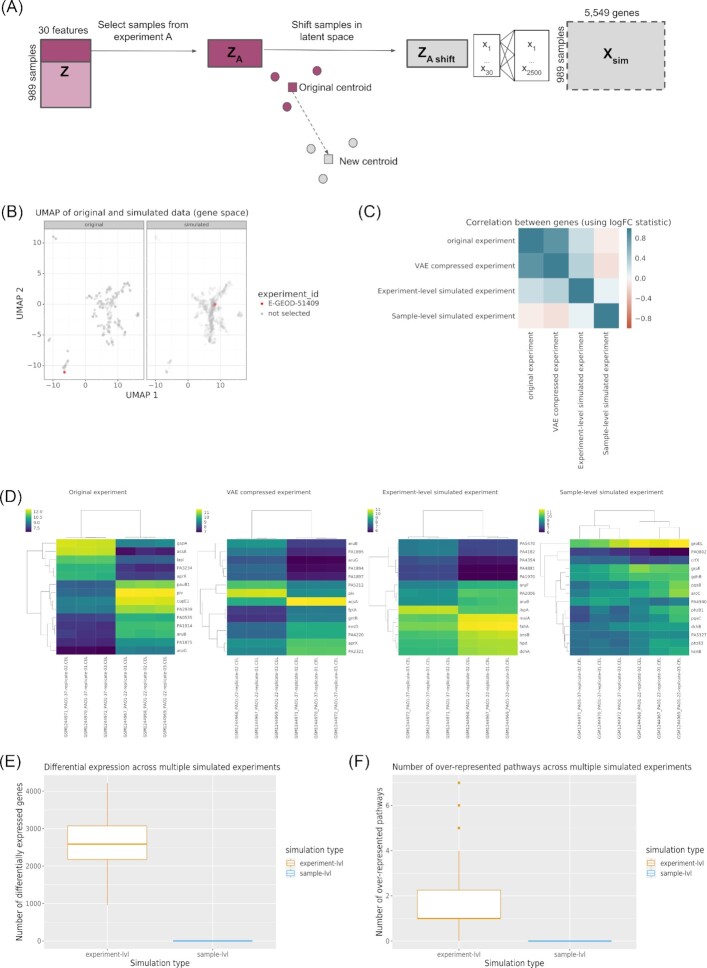
Figure 3:
Simulating gene expression compendia by experiment. (A) Workflow to simulate gene expression per experiment. (B) UMAP projection of P. aeruginosa gene expression data highlighting a single experiment, E-GEOD-51409 (red), in the original dataset (left) and the simulated dataset (right), which was subsampled to 1,000 samples. (C) Correlation matrix showing the correlation between genes, based on their logFC, in the original experiment (E-GEOD-51409) vs VAE compressed experiment; original experiment vs simulated experiment using the original experiment as a template (experiment-level simulated); original vs random simulated (sample-level simulated) experiment. (D) Differential expression analysis of experiment E-GEOD-51409 (left), VAE-compressed experiment (middle-left), random simulated experiment (middle-right), and simulated experiment using the original experiment as a template (right). (E) Number of differentially expressed genes identified across 100 simulated experiments generated using experiment-level simulation and sample-level simulation. (F) Number of enriched pathways identified across 100 simulated experiments generated using experiment-level simulation and sample-level simulation. Circles denote outliers.logFC: log fold-change.