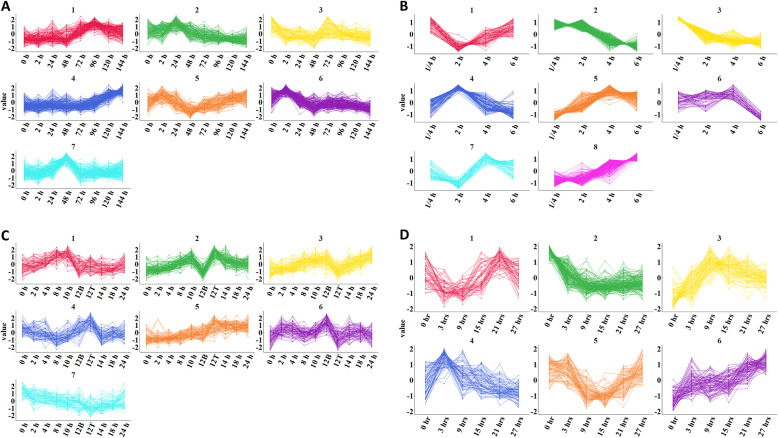
Fig. 8.
K-means clustering of transcriptomic data from wild type for genes in the phenotype dataset. The expression values for each gene in the phenotype dataset were standardized to have a mean of 0 and a standard deviation of 2. Genes were partitioned into the indicated number of expression profiles using K-means clustering. a. RNA-seq data from a sexual cycle time course. RNA-seq data for eight time points were obtained from Ref. [52]. Genes were partitioned into seven expression profiles. b. RNA-seq data from a time course of conidial germination. RNA-seq data for four time points were obtained from Ref. [53]. Genes were partitioned into eight expression clusters. c. Microarray data from a time course of conidiation. Microarray data for 10 time points were obtained from Ref. [54]. Genes were partitioned into seven expression clusters. d. Microarray data during a colony development time course. Microarray data for six time points were obtained from Ref. [55]. Genes were partitioned into six expression clusters