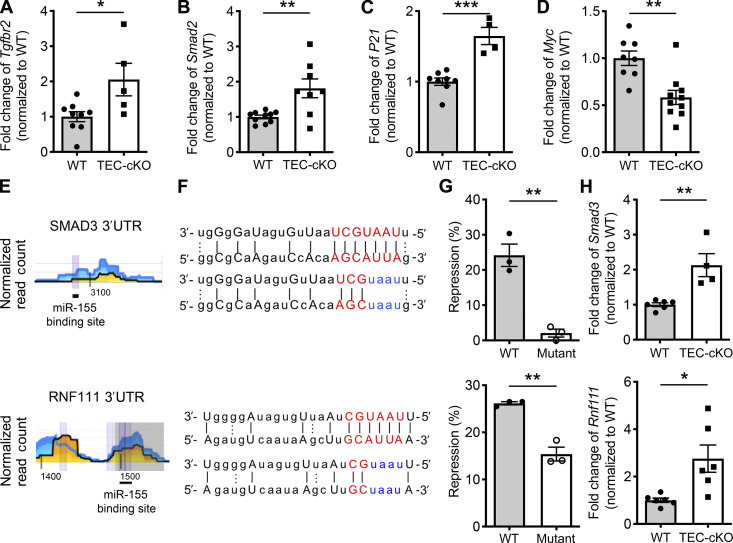
Figure 4.
miR-155 regulates TGFβ signaling in mature mTECs via targeting multiple key components. (A–D) qPCR analyses for the expressions of Tgfbr2 (A), Smad2 (B), P21 (C), and Myc (D) in sorted CD80hiMHCIIhi mature mTECs from 5–6-wk-old WT and TEC-cKO mice. The n-fold changes on the basis of each corresponding WT controls were shown. (E and F) High-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation analyses (the underlying numbers represent the nucleotide position related to the start of the 3′UTR (E) and sequence alignments (F) of the putative miR-155 binding sites in 3′UTR of SMAD3 (upper) and RNF111 (lower). Mutations of the corresponding miR-155 target sites are shown in blue. (G) Ratios of repressed luciferase activity of cells with SMAD3 3′UTR (upper) and RNF111 3′UTR (lower) with or without mutations in the seed sequences in the presence of miR-155 compared with cells transfected with empty vector. (H) qPCR analyses of the expressions of Smad3 (upper) and Rnf111 (lower) in sorted CD80hiMHCIIhi mature mTECs from 5–6-wk-old WT and TEC-cKO mice. Each symbol represents an individual mouse, and the bar represents the mean. Data are pooled from at least three independent experiments. Results of Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.