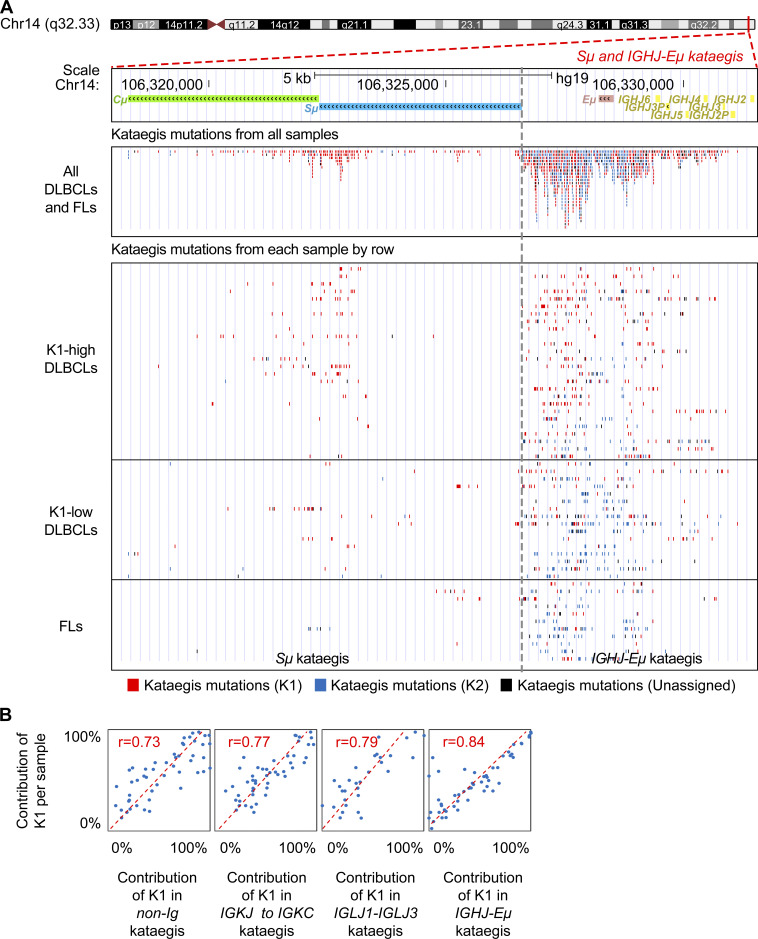
Figure S2.
The overall contribution of K1 for a given sample is highly correlated to its contribution to the different regions within the same sample, except S regions. (A) The mutation pattern and distribution of kataegis in the Sμ and IGHJ-Eμ regions. Top: For the region of interest, the gene structure and location on the chromosome are shown. The kataegis mutations are shown together for all samples (middle) or separately by each sample (bottom). Each dot represents a kataegis mutation. The red and blue dots represent mutations belonging to K1 and K2, respectively. The black dots represent unclassified mutations to either K1 or K2. For the bottom panel, each row represents a sample. The samples are in the same order as in Fig. 2 F and are grouped as K1-high DLBCL, K1-low DLBCL, and FL. Mutations are grouped in either the Sμ region (left) or IGHJ-Eμ region (right). The kataegis across both regions were initially identified together due to their closely adjacent positions in the genome. To reveal the characteristics of kataegis for both parts, kataegis located in the Sμ and IGHJ-Eμ regions were divided for further analysis. For the divided kataegis, only those with at least 10 mutations were counted for further analysis. (B) The contribution of K1 for a given sample highly correlated to its contribution to different regions, including the most targeted IGHJ-Eμ, IGLJ1-IGLJ3, and IGKJ to IGKC regions, as well as the non-Ig regions. Correlation was calculated by the PCC.