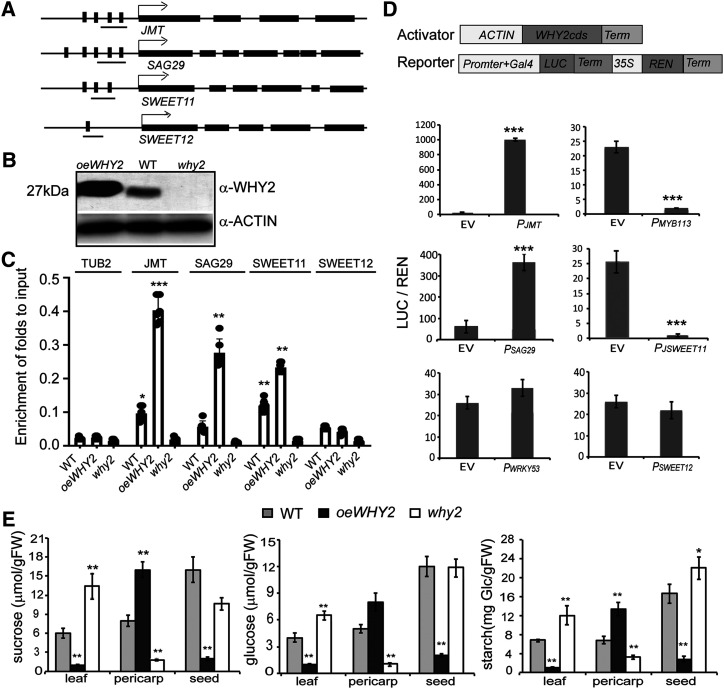
Figure 7.
WHY2 binds to upstream regions of JMT, SAG29, and SWEET11 and alters their expression. A, Diagram of the promoters of JMT, SAG29, SWEET11, and SWEET12 genes. B, Western blot detection of WHY2 protein in the rosette of 7-week-old oeWHY2, why2, and wild-type (WT) plants. C, Chromatin immunoprecipitation (ChIP) assay in rosette leaves of 7-week-old Arabidopsis oeWHY2, why2, and wild-type plants. Antibody against WHY2 peptide was used. TUB2 was used as a negative control. The fold enrichment of ChIP is relative to input. Data represent mean ± sd of five biological replicates. Asterisks denote statistically significant differences from the enrichment of TUB2, calculated using Student’s t test: (*P < 0.05; **P < 0.01, ***P < 0.001). D, LUC/REN dual activation assay, as above. Agrobacterium cells containing the vectors expressing WHY2-FLAG (ACTIN:WHY2-FLAG) or vectors expressing candidate promoter fragments plus GAL4: LUC-REN were coinjected into Nicotiana benthamiana leaves. The WRKY53 promoter was used as a negative control. E, Suc, Glc, and starch content in the leaf, pericarp, and seeds of 7-week-old oeWHY2, why2, and wild-type plants. Shown are mean ± se of six biological replicates. Asterisks denote statistically significant differences from the empty vector or wild type calculated using Student’s t test: *P < 0.05; **P < 0.01; and ***P < 0.001.