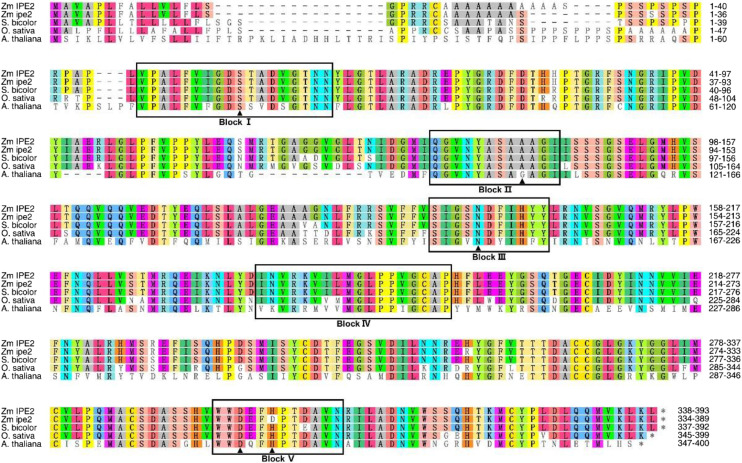
Figure 7.
Amino acid alignment of IPE2 homologs in maize, sorghum, rice, and Arabidopsis. Black boxes indicate the five conserved blocks. Amino acids marked by the solid black triangle indicate the putative active sites. Among them, S in Block I and D and H in Block V indicate the catalytic triad; and S in Block I, A/G in Block II, and N in Block III indicate the oxyanion hole. Accession numbers for homologs in sorghum, rice, and Arabidopsis are XP 002451978.1, XP_015623170.1, and NP_567372.1, respectively.