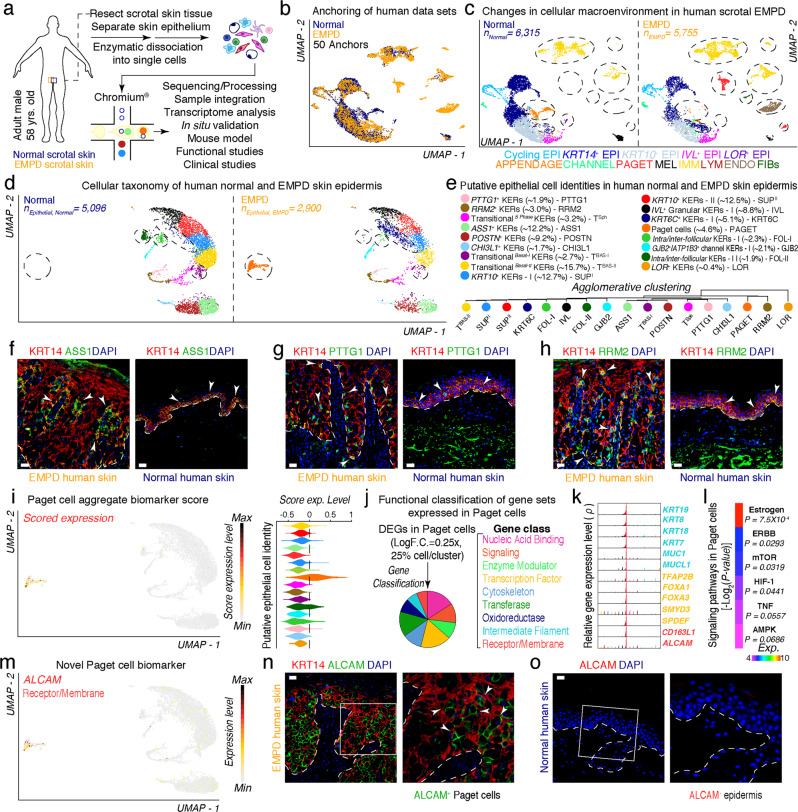
Fig. 1. Characterization of the epithelial macroenvironment of EMPD skin using 3′-end scRNA-seq.
a Schematic of patient anatomy, cell isolation, processing, and capture of single cells by droplet-based device, 3′-end scRNA-seq, and downstream analyses. b Anchoring (50 anchors) of normal (n = 6315 viable cells) and EMPD (n = 5755 viable cells) data sets visualized in UMAP space. c Clustering and neighbor identification of anchored data sets split by condition. Thirteen putative cell communities were identified and color coded. Putative community identity identified by bona fide biomarker expression is defined, shown in Supplementary information, Fig. S3. d Epithelial cellular taxonomy of human normal (n = 5096 epithelial cells) and EMPD (n = 2900 epithelial cells) skin epidermis split by condition. Epithelial cells that appear to be condition-specific are demarcated with broken oval shapes. e Putative epithelial cell identity identified by bona fide marker expression with their respective proportions is defined. Agglomerative clustering demonstrates hierarchical relationships between epithelial cells. f–h Immunofluorescence for ASS1/KRT14, PTTG1/KRT14 and RRM2/KRT14 in EMPD and normal skin. White arrows point to ASS1+, PTTG1+, RRM2+ cells. i Paget cells were identified by implementation of a ‘Paget cell aggregate biomarker score’ defined by a core set 10 PD-associated biomarkers. These PD biomarkers included: KRT8, AR, PDPN, GATA3, MYD88, MUC1, KRT7, VIM, MMP9, and PIP. Scored cells were projected on UMAP, evaluated as feature plot and quantified using violin plot. Gene expression levels are color coded. j Functional classification of select gene sets expressed in Paget cells (Log F.C. = 0.25×, 25% cell/cluster, Wilcox test, P < 0.05). Selected gene sets were plotted using a pie chart and gene classes were color coded. k High-density plots show distribution of select Paget cell-specific transcripts including known and novel biomarkers. l Heatmap showing signaling pathways activated in Paget cells. These pathways included Estrogen (P = 4.75 × 10−3), ERBB (P = 0.0293), mTOR (P = 0.0319), HIF-1 (P = 0.0441), TNF (P = 0.0557), AMPK (P = 0.0686). m Distribution of ALCAM expression overlaid on feature plot shows high and unique expression in Paget cells. Immunofluorescence of KRT14 and ALCAM in EMPD skin (n) and human normal skin (o). Insets represent magnified areas. Representative images are shown. Epidermis and dermis are demarcated with broken line. Scale bars, 25 μm (f–h, n, o).