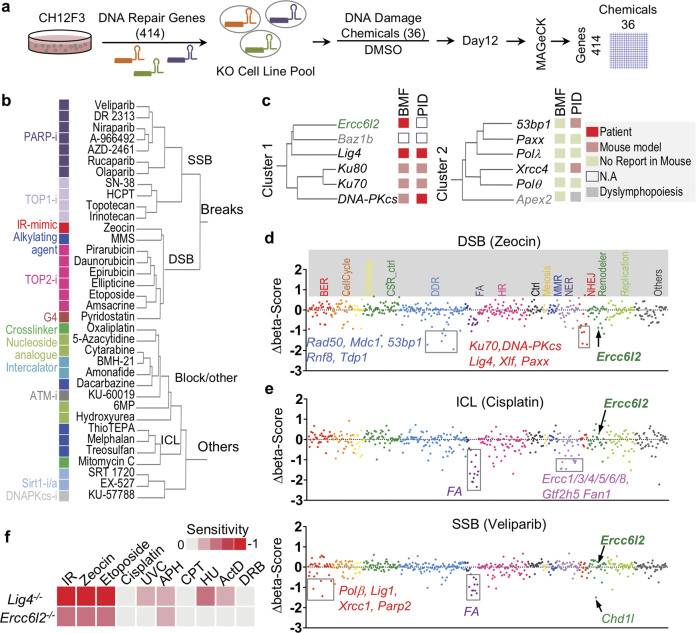
Fig. 1. CRISPR-chemical screening uncovers ERCC6L2 as a potential NHEJ factor.
a Schematic illustration of chemical genetic screening procedure. b Clustering tree of chemicals. Chemical names are listed and annotated by colored blocks. Similar chemicals based on their inhibitory targets or caused DNA damages types are marked with same color. c Tree-view of two NHEJ gene clusters. Gene names are listed. Roles in bone marrow failure (BMF) or primary immunodeficiency (PID) are annotated by colored blocks. d, e Sensitivity of gene knockouts to Zeocin (d) or Cisplatin/Veliparib (e) treatment. Genes are grouped by gene ontology and depicted with different colors based on the involved biological processes along the x-axis. The beta-score difference (chemical-DMSO) indicating positive or negative enrichment in the chemical treatment samples was calculated from two replicates and is plotted for each gene. Representative negatively enriched genes are labeled. FA indicated Fanconi anemia genes. f Sensitivity of ERCC6L2-deficient or LIG4-deficient B cells to different treatments. Cell viability curve was calculated and the area-under-the-curve (AUC) was computed. Heat map of sensitivity, which is indicated as “log2(AUCKO/AUCWT)”, is plotted. IR γ-irradiation, UVC ultraviolet wavelength 254 nm, APH aphidicolin, CPT camptothecin, HU hydroxyurea, ActD actinomycin D, DRB 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside.