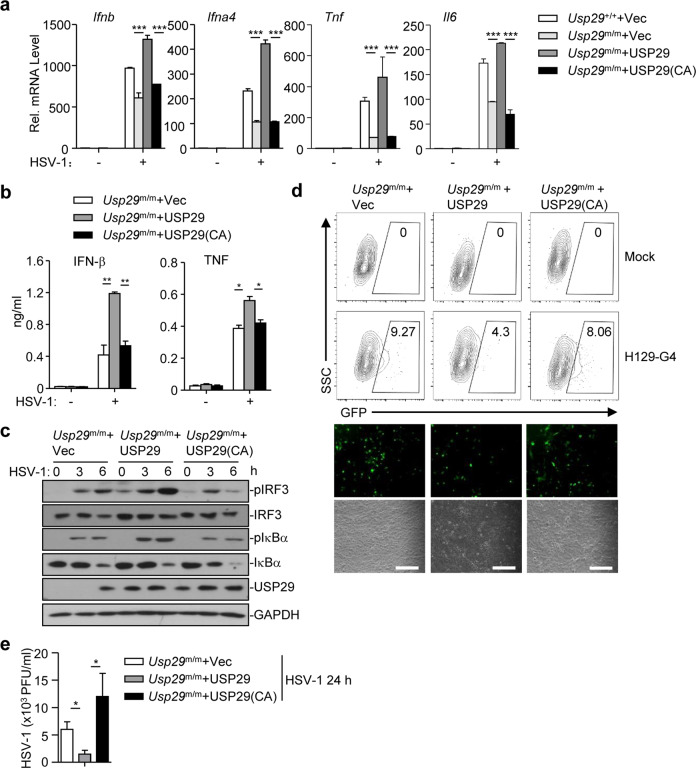
Fig. 5. The DUB activity of USP29 is required for optimal cellular antiviral responses.
a qRT-PCR analysis of Ifnb, Ifna4, Tnf, or Il6 mRNA in Usp29+/+ MLFs reconstituted with the empty vector (Usp29+/++Vec) or in Usp29m/m MLFs reconstituted with the empty vector (Usp29m/m + Vec), USP29 (Usp29m/m + USP29), or USP29 (C298A) [Usp29m/m + USP29 (C298A)] infected with HSV-1 for 0–6 h. b, c ELISA analysis of IFN-β and TNF (b) and immunoblot analysis of phosphorylated and total IRF3 and IκBα, USP29, and GAPDH (c) in Usp29m/m MLFs reconstituted with the empty vector (Usp29m/m + Vec), USP29 (Usp29m/m + USP29), or USP29 (C298A)[Usp29m/m + USP29(C298A)] infected with HSV-1 for 0–6 h. d Flow cytometry analysis (upper flow charts), fluorescent microscopy imaging (lower images) of Usp29m/m MLFs reconstituted with the empty vector (Usp29m/m + Vec), USP29 (Usp29m/m + USP29), or USP29 (C298A) [Usp29m/m + USP29(C298A)] infected with or without H129-G4 for 24 h. e Plaque assays of Usp29m/m MLFs reconstituted with the empty vector (Usp29m/m + Vec), USP29 (Usp29m/m + USP29), or USP29 (C298A) [Usp29m/m + USP29(C298A)] infected with or without HSV-1 for 24 h. *P < 0.05; **P < 0.001; ***P < 0.001 (analysis of two-way ANOVA followed by Bonferroni post-test). Scale bars represent 300 μm. Data are representative of three (a, b, d, e) or two (c) independent experiments (means ± SD in a, b, e).