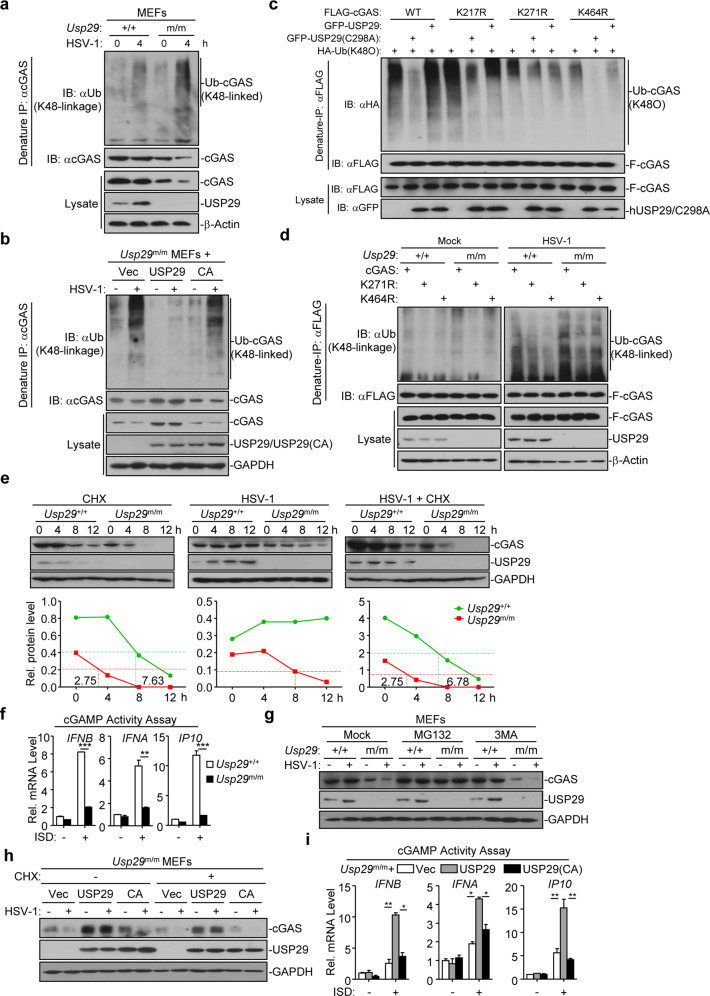
Fig. 6. USP29 deubiquitinates and stabilizes cGAS.
a Denature-IP (with anti-cGAS) and immunoblot analysis [with anti-Ub (K48-linkage), anti-cGAS, anti-USP29 or anti-β-Actin] of Usp29+/+ and Usp29m/m MEFs left uninfected or infected with HSV-1 for 4 h. b Denature-IP (with anti-cGAS) and immunoblot analysis [with anti-Ub (K48-linkage), anti- cGAS, anti-USP29 or anti-GAPDH] of Usp29m/m MEFs reconstituted with empty vector, USP29 or USP29(C298A) left uninfected or infected with HSV-1 for 4 h. c Denature-IP (with anti-FLAG) and immunoblot analysis (with anti-FLAG, anti-HA or anti-GFP) of HEK293 cells transfected with plasmids encoding HA-Ub(K48O), FLAG-cGAS or FLAG-cGAS mutants together with an empty vector, GFP-USP29 or GFP-USP29(C298A) for 24 h. d Denature-IP (with anti-FLAG) and immunoblot analysis [with anti-Ub (K48-linkage), anti-FLAG, anti-USP29 or anti-β-Actin] of Usp29+/+ and Usp29m/m MLFs reconstituted with cGAS, cGAS(K271R) or cGAS(K464R) left uninfected or infected with HSV-1 for 6 h. e Immunoblot analysis of cGAS, USP29 and GAPDH (upper blots) and quantitation of the intensities of cGAS (relative to GAPDH) (lower graphs) in Usp29+/+ and Usp29m/m MLFs infected with or without HSV-1 in the presence or absence of CHX for 0–12 h. f Usp29+/+ and Usp29m/m MLFs were transfected with ISD45 for 4 h, and then the cell extracts containing cGAMP were delivered to digitonin-permeabilized HFFs for 4 h followed by qRT-PCR analysis. g Immunoblot analysis of cGAS, USP29 and GAPDH in Usp29+/+ and Usp29m/m MEFs that were treated with MG132 or 3MA, followed by HSV-1 infection for 0–4 h. h Immunoblot analysis of cGAS, USP29 and GAPDH in Usp29m/m MEFs reconstituted with the empty vector (Vec), USP29 (USP29), or USP29 (C298A) (CA) that were infected by HSV-1 in the presence or absence of CHX for 0–4 h. i Usp29m/m MEFs reconstituted with the empty vector (Vec), USP29 (USP29) or USP29 (C298A) (CA) were transfected with ISD45 for 4 h, and then cell extracts containing cGAMP were delivered to digitonin-permeabilized HFFs for 4 h followed by qRT-PCR analysis. *P < 0.05; **P < 0.001; ***P < 0.001 (analysis of two-way ANOVA followed by Bonferroni post-test or two-tailed Student’s t-test). Data are representative of three (f, i) or two (a, b, c, d, e, g, h) independent experiments (means ± SD in f, i).