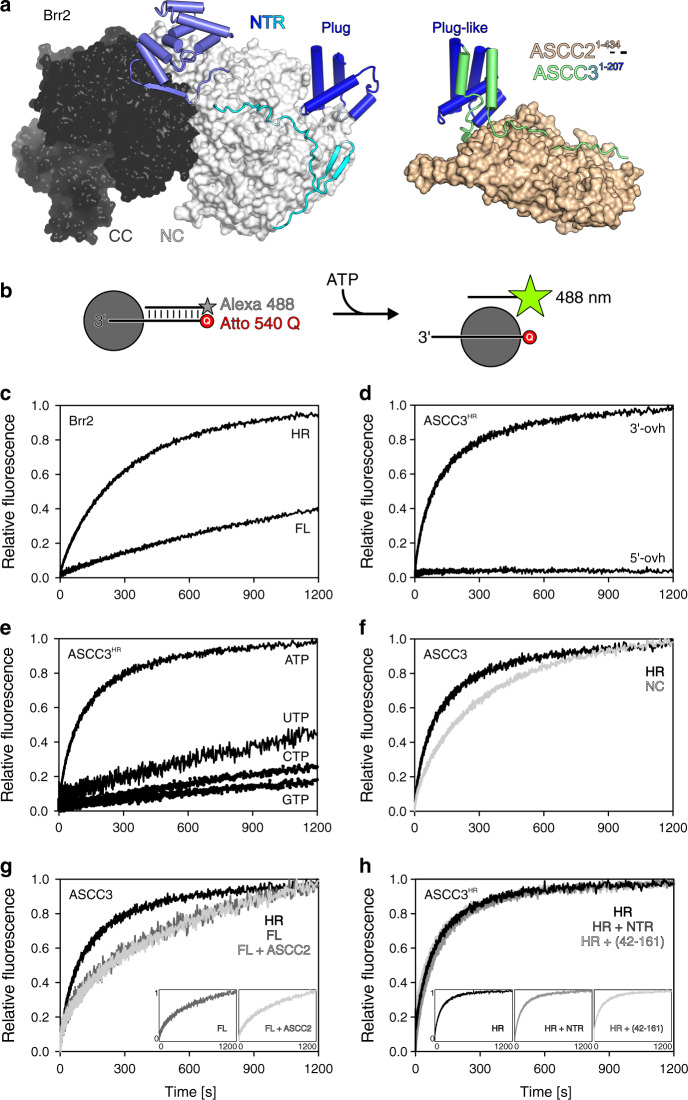
Fig. 5. Unwinding assays.
a Comparison of the structure of full-length yeast Brr2 (left) and the present ASCC21–434-ASCC31–207 complex (right) after superposition of the plug/plug-like domains (blue). Human Brr2 in the U4/U6•U5 tri-snRNP or in the pre-catalytic spliceosome exhibits a plug domain very similar to yeast Brr2 blocking RNA access42,43 (PDB IDs 3JCR, 6QW6, 6QX9). NC, N-terminal cassette; CC, C-terminal helicase cassette. b Experimental setup for stopped-flow/fluorescence-based unwinding assays. Gray sphere, helicase; star symbol, fluorophore (Alexa 488); Q, quencher (Atto 540 Q). c Stopped-flow/fluorescence-based assays monitoring unwinding of a 3’-overhang RNA by Brr2FL or Brr2HR, showing that auto-inhibition, which has been documented for Brr2 using gel-based unwinding assays33, can be readily detected using the present experimental setup. d Stopped-flow/fluorescence-based assays monitoring unwinding of DNA bearing a 3’-overhang (3’-ovh), and lack of unwinding of a 5’-overhang (5’-ovh) DNA by ASCC3HR upon addition of ATP. e Nucleotide preference of ASCC3HR in unwinding a 3’-overhang DNA. f Stopped-flow/fluorescence-based assays monitoring unwinding of 3’-overhang DNA by ASCC3NC compared to ASCC3HR using ATP. g Stopped-flow/fluorescence-based assays monitoring unwinding of a 3’-overhang DNA by the indicated ASCC3 constructs in the absence or presence of ASCC2 using ATP. Insets, side-by-side presentation of the largely overlapping curves. h Stopped-flow/fluorescence-based assays monitoring unwinding of a 3’-overhang DNA by ASCC3HR alone or in the presence of ASCC3NTR or ASCC342–161 using ATP. Insets, side-by-side presentation of the largely overlapping curves. FL/HR/NC/NTR/(42–161), ASCC3 variants as defined in the text.