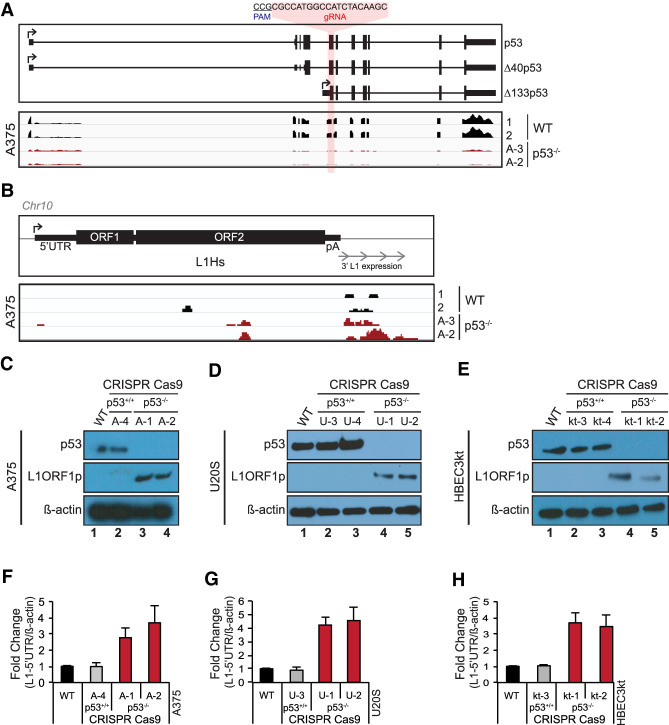
Figure 1.
p53 loss provokes human L1 expression. The p53 gene was mutated using a CRISPR guide RNA targeting the first common exon found in all annotated p53 isoforms, as illustrated in the top panel of A. In A and B, two independently edited p53−/− lines, designated A-3 and A-2, were analyzed. In C and F, independently edited p53−/− lines A-1 and A-2 were analyzed. In the bottom panel of A, normalized RNA sequencing expression levels from wild-type (WT; black) and p53−/− A375 cells (red) are shown. (B) Normalized expression of readthrough transcription from a single, uniquely identifiable L1Hs element in wild-type and p53−/− A375 cells. Western blot in WT, cas9-treated control and p53−/− A375 human melanoma (C), U2OS osteosarcoma (D), and HBEC3kt immortalized normal human lung (E) cells lines for p53 (top panels) and L1ORF1p (middle panels). (Bottom panels) β-Actin is presented as a loading control. Normalized L1-5′UTR mRNA levels in WT, cas9-treated control and two independent p53−/− A375 (F), U2OS (G), and HBEC3kt (H) cell lines assessed by ddPCR. L1-5′UTR transcript levels were normalized to β-actin and relative fold change was calculated with respect to parental wild type. Bar graphs are the average of three independent experiments (n = 3) with error bars representing the standard deviation. p53−/− samples were significantly different from parental wild type (two-tailed t-test P-value < 0.05) and cas9-treated controls (P-value < 0.05).