Figure 4.
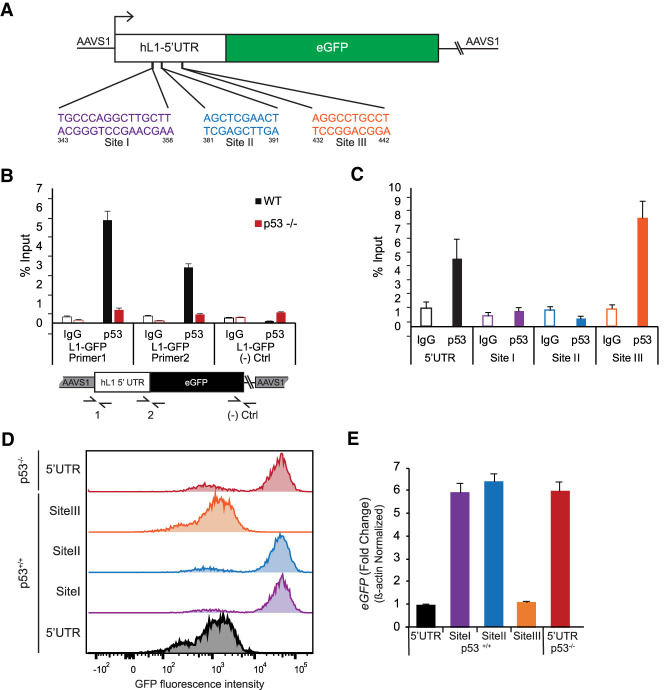
p53 physically binds to repress the L1-5′UTR The schematic in A shows empirically and computationally defined p53 binding sites in the 5′UTR of L1-5′UTR-eGFP. Note that site I, site II, and site III were independently targeted for deletion. (B) ChIP for p53 and control IgG performed with illustrated primer pairs (1 and 2) that uniquely detect binding at the L1 reporter. Note the absence of signal in p53−/− cells and at the L1-GFP negative control region spanning vector-derived sequences (Ctrl primer pair). Error bars represent 95% confidence intervals. (C) ChIP testing of p53-binding site deletions indicated in A along with the intact wild-type 5′UTR interval. Note, site I and site II contribute to p53 binding, but site III had no effect. Error bars represent 95% confidence intervals. In D, flow cytometry was used to detect for GFP intensities in the intact and mutant L1-5′UTR-eGFP reporters in A375 cells. Note that sites I and site II are required for p53 repression of the L1 expression reporter (see Supplemental Fig. S4L for another biological replicate). (E) Shows that corresponding eGFP mRNA transcript levels mirror GFP intensities seen in these same cells. Bar graphs are averages of three biological replicates. Error bars represent standard error of the mean.