Figure 4.
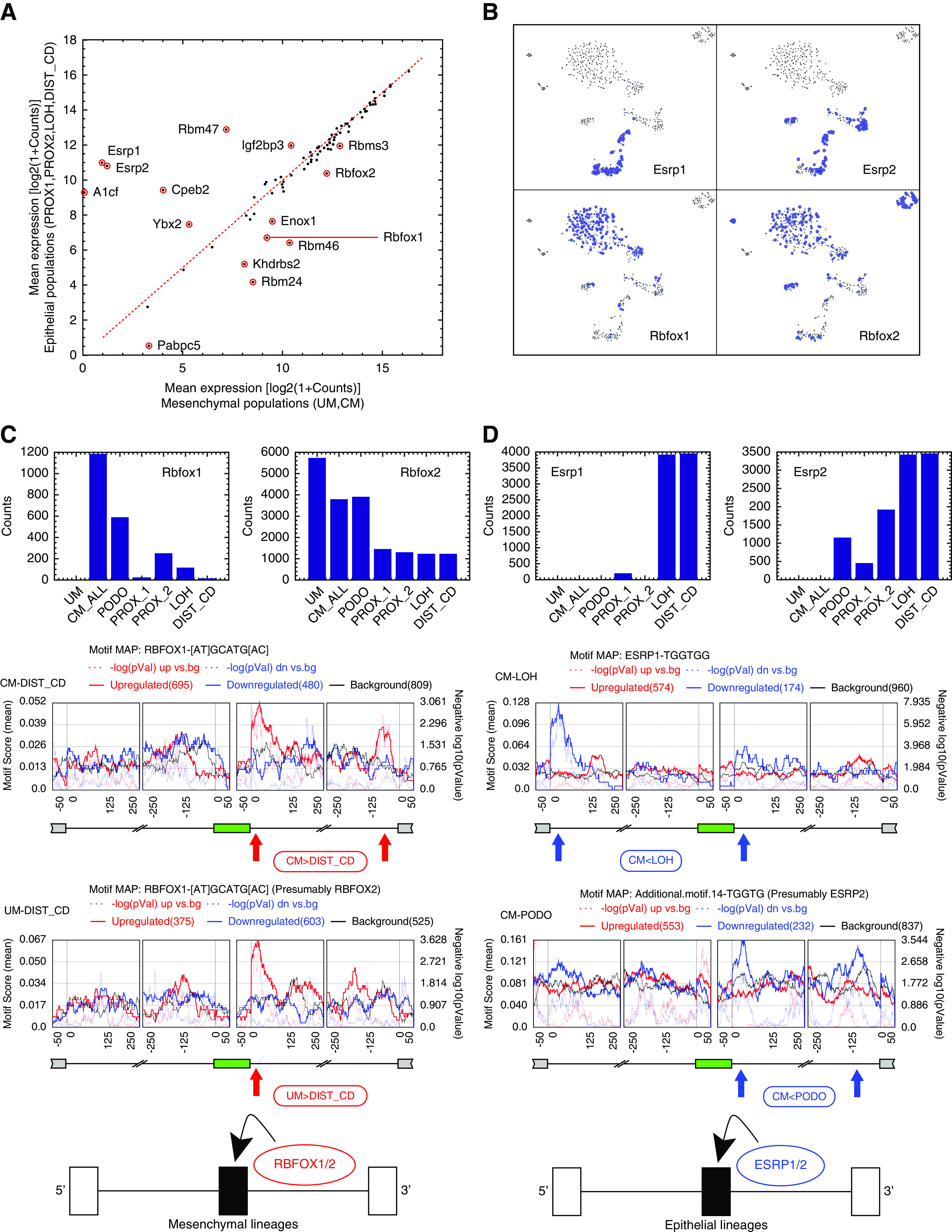
Differential expression of RNA binding proteins and RNA binding motif enrichment analysis suggest that Esrp1/2 and Rbfox1/2 are splicing regulators of the Mesenchymal to Epithelial transition (MET) that occurs during kidney development. (A) Shown is a comparison between mesenchymal and epithelial states of the mean expression levels of 84 RNA binding proteins (RBPs) that are known from the literature to regulate splicing through binding of mRNA transcripts. We used expression levels from bulk in silico transcriptomes that represent each cell population. (B) A tSNE plot of the single-cell profiles, highlighting cells that express the putative splicing regulators Rbfox1/2 and Esrp1/2. It can be seen that Rbfox1 and Rbfox2 are highly expressed in the mesenchymal populations, whereas Esrp1 and Esrp2 are highly expressed in the epithelial populations. The area of each circle is proportional to log2(1+expression) of each gene in that particular cell. (C and D) Cassette exons that are over-expressed in the mesenchymal populations contain a significant enrichment of Rbfox1/2 binding motifs at their downstream introns.12,40,66 Likewise, cassette exons that are over-expressed in the epithelial populations contain a significant enrichment of Esrp1/2 binding motifs in their downstream introns,19,42,43 and in some cases (CM versus LOH), also in the far 5′ end of their upstream introns (as previously observed in EMT42). This indicates that Rbfox1/2 and Esrp1/2 are splicing regulators12 involved in the MET that occurs during kidney development. The annotations “CM_ALL” and “CM” are used interchangeably to represent all cells that were classified as belonging to the CM (see also Methods and Supplemental Appendix 1).
