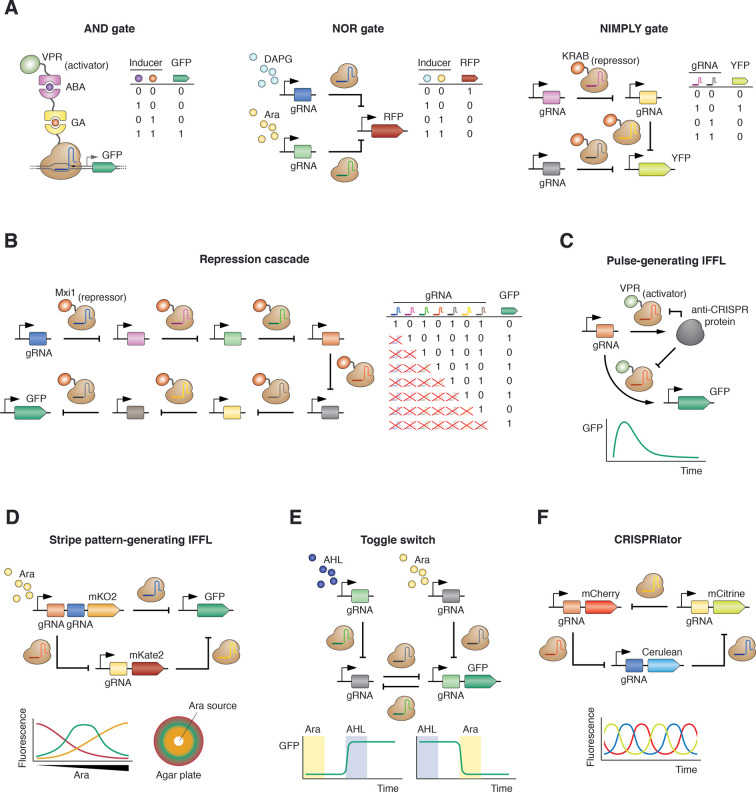
Figure 2. CRISPR-based synthetic gene circuits.
(A) Examples of CRISPR-based logic gates. Left: AND gate based on ligand-induced heterodimerization domains implemented in human cells [105]. Only in the presence of the two ligands can the activator domain be recruited to the DNA-bound dCas9 and activate GFP expression. Middle: NOR gate constructed in E. coli using gRNAs expressed from inducible promoters [41]. Both gRNAs bind to and repress RFP, and thus only in the absence of both inducers can RFP be expressed. Right: NIMPLY gate in human cells mediated by KRAB-fused dCas9 [142]. YFP expression is enabled when one of the input gRNAs (but not the other) is present. (B) Seven-layer repression cascade built in yeast by using dCas9-Mxi1 [71]. Each new layer inverts the ‘sign’ (i.e. 1 or 0, high or low) of the downstream layers and of the final output. (C) Pulse-generating IFFL in human cells by leveraging dCas9-VPR inactivation by anti-CRISPR proteins [137]. The result is a temporal peak of gene expression. (D) Stripe-forming IFFL in E. coli [69]. In the presence of a gradient of arabinose (Ara, the input inducer), the dCas9-governed circuit forms a peak (stripe) of gene expression at intermediate Ara concentrations. Genetically identical bacteria carrying this circuit form a 3-color pattern in an agar plate when subjected to an Ara gradient. (E) dCas9-controlled toggle switch in E. coli [69]. The circuit can be toggled between the two stable states (green and non-green) through the addition of inducers to the medium: after inducing one of the states, the cells remain in that state even in the absence of any inducer, and the same is true for the opposite state. (F) The CRISPRlator, the first CRISPR oscillator [69]. A dCas9-mediated closed-ring repression topology generates periodic oscillations in E. coli cells.