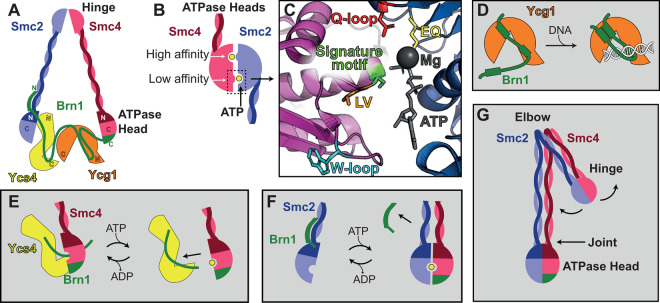
Figure 1. Condensin conformational changes.
(A) Schematic of the subunits that make up S. cerevisiae condensin, indicating the hinge dimerisation domain, and ATPase head domains. (B) Schematic of ATP bound ATPase head domain, indicating high and low affinity ATPase sites in Smc4 and Smc2, respectively. (C) An atomic model of engaged Smc2 and 4 ATPase heads, created by superimposition of C. thermophilum Smc2 and 4 crystal structures (6QJ0 and 6QJ2) [43] with C. thermophilum/S. cerevisiae chimeric engaged Smc1/3 heads (6QPW) [50]. Commonly used mutations to prevent ATP binding (Q-loop, red), prevent head dimerisation (signature motif, green) and slow ATP hydrolysis (EQ, yellow) are indicated, as well as W-loop (cyan) important for binding Ycs4 and the LV mutation (orange) which affects ATP hydrolysis if present in Smc2 and Z-loop formation if present in Smc4. (D) Brn1 creates a safety belt, locking DNA to the Ycg1 HAWK subunit. (E) Ycs4/Brn1 binds Smc4/Brn1, but ATP interferes with this interaction, creating a possible ATP dependent cycle. (F) Brn1 N-terminus binding to Smc2 is lost in the presence of ATP dependent head dimerisation, creating another possible ATP dependent cycle. (G) SMC dimers are able to bend at the elbow region such that the hinge moves towards the ATPase heads. SMC coiled-coils also have some flexibility at the joint region, near the ATPases heads.