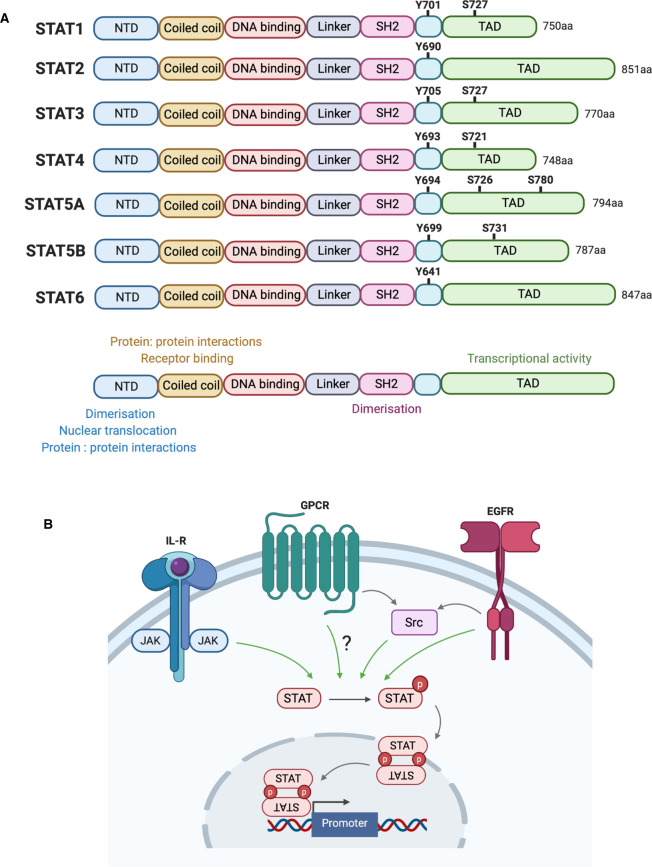
Figure 2. Introduction to the STAT family of transcription factors.
(A) Homologous STAT family domain structure. All family members contain an N terminal domain (NTD) that is involved in dimerisation, nuclear translocation and interaction with other proteins; a coiled coil domain involved in protein: protein interactions and receptor binding; a DNA binding domain (DBD) that directly binds to target DNA sequences; a linker region; an SH2 domain involved in dimerisation by interacting with pTyr of another activated monomer and a C-terminal transactivation domain (TAD) that regulates STAT transcriptional activity. Total amino acid length of each STAT member is listed. A conserved tyrosine residue is shared between all family members, corresponding to Tyr701 in STAT1. Additionally, some family members share a conserved serine residue in the TAD domain, corresponding to Ser727 in STAT1. (B) STAT activation mechanisms. STAT members can be tyrosine phosphorylated by cytoplasmic JAK kinases activated at cytokine receptors, in addition to GPCR activated non-receptor tyrosine kinases like Src and growth factor receptors such as EGFR. Phosphorylation (represented by red circles) allows for reciprocal interactions between pTyr and SH2 domains of monomers that allow dimer formation. Translocation of active dimers to the nucleus allows for binding upstream of target gene promoters.