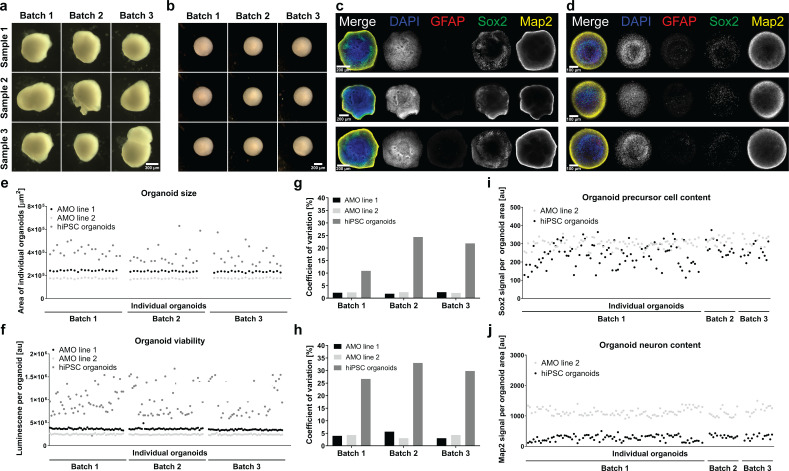
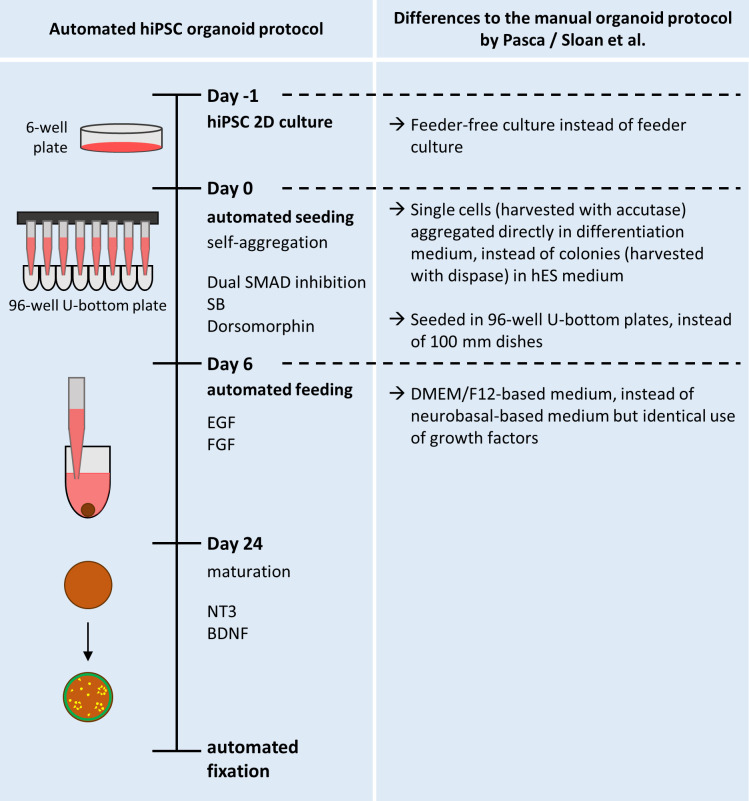
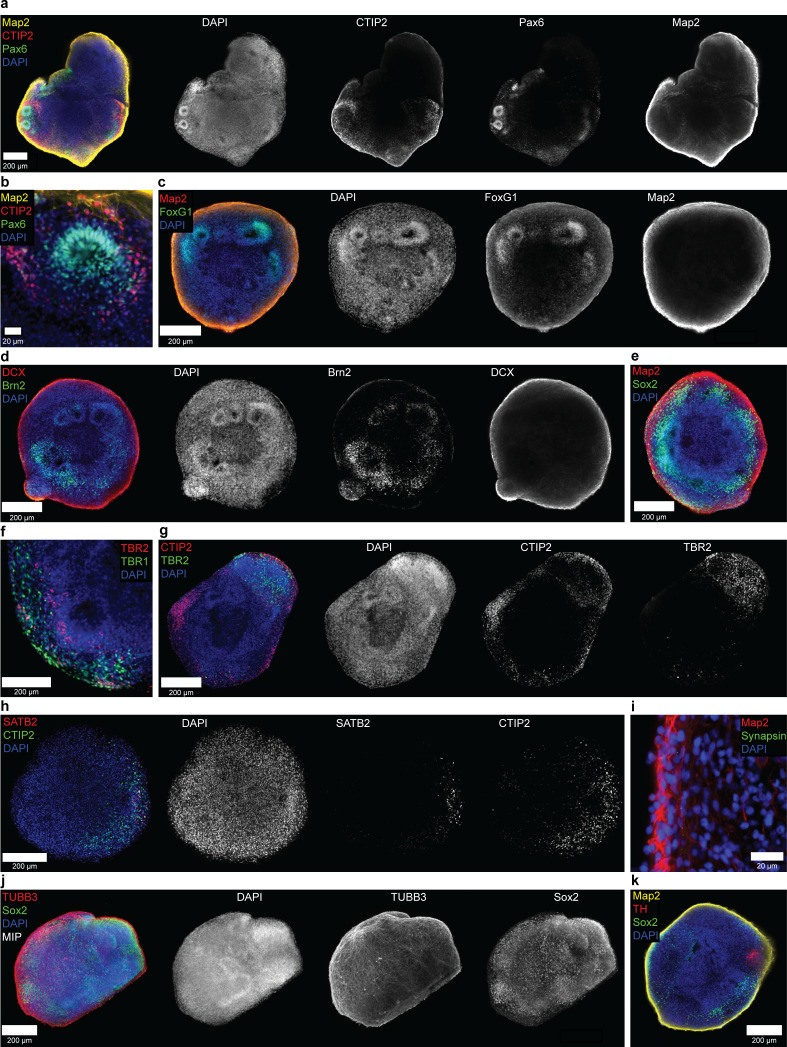
Figure 7. smNPC-derived AMOs are morphologically, structurally, and functionally more homogeneous than automated hiPSC-derived organoids.
(a/b) Light microscopy images of hiPSC-derived organoids (a) and AMOs (b) generated from the same cell line demonstrating the higher morphological homogeneity of AMOs at day 30 of differentiation.(c/d) Single optical confocal slices of either hiPSC-derived organoids (c) or AMOs (d) at day 30 stained for DAPI, the astrocyte marker GFAP, the neural precursor marker Sox2, and the neuronal marker Map2. The direct comparison illustrates the higher level of structural homogeneity as well as accelerated maturation, especially the earlier emergence of GFAP+ astrocytes in AMOs. Rows depict three samples from one batch. (e/f) Size (area of the largest cross section) and cell viability measurements of individual organoids from three independent batches (per cell line/differentiation protocol) illustrating the high homogeneity of AMOs compared to standard hiPSC organoids. (g/h) Coefficients of variation calculated based on the data shown in (e) and (f). (i/j) Quantitative whole mount staining (see also Figure 6) for Sox2 (i), and Map2 (j) showing the higher variability of hiPSC organoids compared to AMOs from the same iPSC line even after normalization to the organoid area. All data gathered from organoids at day 30 of differentiation. Scale bars: 300 μm (a), 200 μm (b/c), 100 μm (d). Also see Figure 7—figure supplements 1 and 2.